Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
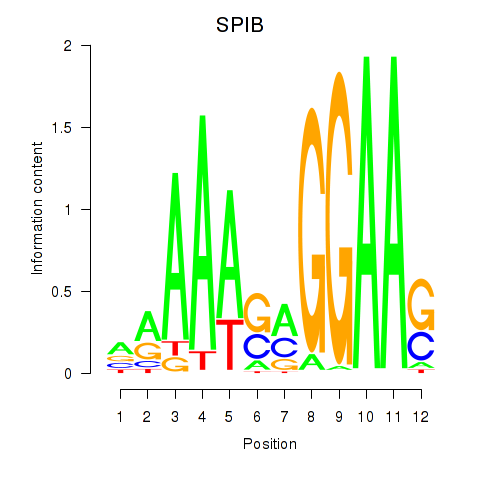
Results for SPIB
Z-value: 3.64
Transcription factors associated with SPIB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPIB
|
ENSG00000269404.2 | SPIB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SPIB | hg19_v2_chr19_+_50922187_50922215 | 0.84 | 4.5e-05 | Click! |
Activity profile of SPIB motif
Sorted Z-values of SPIB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SPIB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_15114492 | 20.45 |
ENST00000541546.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_-_160681593 | 16.29 |
ENST00000368045.3 ENST00000368046.3 |
CD48 |
CD48 molecule |
| chr10_-_121302195 | 12.14 |
ENST00000369103.2 |
RGS10 |
regulator of G-protein signaling 10 |
| chr12_-_15114603 | 11.91 |
ENST00000228945.4 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_-_31230650 | 11.72 |
ENST00000294507.3 |
LAPTM5 |
lysosomal protein transmembrane 5 |
| chr10_-_98031310 | 10.49 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr10_-_98031265 | 9.96 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr3_-_121379739 | 9.07 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr16_+_30194916 | 8.92 |
ENST00000570045.1 ENST00000565497.1 ENST00000570244.1 |
CORO1A |
coronin, actin binding protein, 1A |
| chr2_+_143886877 | 8.79 |
ENST00000295095.6 |
ARHGAP15 |
Rho GTPase activating protein 15 |
| chr17_-_29641104 | 8.77 |
ENST00000577894.1 ENST00000330927.4 |
EVI2B |
ecotropic viral integration site 2B |
| chr5_+_169064245 | 7.77 |
ENST00000256935.8 |
DOCK2 |
dedicator of cytokinesis 2 |
| chr4_-_103266626 | 7.57 |
ENST00000356736.4 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr2_+_68592305 | 7.38 |
ENST00000234313.7 |
PLEK |
pleckstrin |
| chr17_-_29641084 | 7.17 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr1_+_209941827 | 7.11 |
ENST00000367023.1 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
| chr12_+_25205666 | 6.96 |
ENST00000547044.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr1_+_26644441 | 6.81 |
ENST00000374213.2 |
CD52 |
CD52 molecule |
| chr19_+_42381337 | 6.77 |
ENST00000597454.1 ENST00000444740.2 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
| chr1_+_161632937 | 6.53 |
ENST00000236937.9 ENST00000367961.4 ENST00000358671.5 |
FCGR2B |
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chrX_+_78200913 | 6.38 |
ENST00000171757.2 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
| chrX_+_78200829 | 6.23 |
ENST00000544091.1 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
| chr16_+_57392684 | 6.20 |
ENST00000219235.4 |
CCL22 |
chemokine (C-C motif) ligand 22 |
| chr7_+_74188309 | 6.07 |
ENST00000289473.4 ENST00000433458.1 |
NCF1 |
neutrophil cytosolic factor 1 |
| chr12_+_25205568 | 5.99 |
ENST00000548766.1 ENST00000556887.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr22_+_37309662 | 5.99 |
ENST00000403662.3 ENST00000262825.5 |
CSF2RB |
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr6_-_154677900 | 5.95 |
ENST00000265198.4 ENST00000520261.1 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
| chr6_+_391739 | 5.77 |
ENST00000380956.4 |
IRF4 |
interferon regulatory factor 4 |
| chr22_-_37640277 | 5.75 |
ENST00000401529.3 ENST00000249071.6 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr19_+_6772710 | 5.48 |
ENST00000304076.2 ENST00000602142.1 ENST00000596764.1 |
VAV1 |
vav 1 guanine nucleotide exchange factor |
| chr2_+_127413704 | 5.46 |
ENST00000409836.3 |
GYPC |
glycophorin C (Gerbich blood group) |
| chr12_+_25205446 | 5.36 |
ENST00000557489.1 ENST00000354454.3 ENST00000536173.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr2_-_242089677 | 5.29 |
ENST00000405260.1 |
PASK |
PAS domain containing serine/threonine kinase |
| chr2_+_127413677 | 5.27 |
ENST00000356887.7 |
GYPC |
glycophorin C (Gerbich blood group) |
| chr3_+_16926441 | 5.25 |
ENST00000418129.2 ENST00000396755.2 |
PLCL2 |
phospholipase C-like 2 |
| chr22_-_37640456 | 5.23 |
ENST00000405484.1 ENST00000441619.1 ENST00000406508.1 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr4_+_103423055 | 5.18 |
ENST00000505458.1 |
NFKB1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr13_+_31309645 | 5.14 |
ENST00000380490.3 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
| chr3_+_121796697 | 5.06 |
ENST00000482356.1 ENST00000393627.2 |
CD86 |
CD86 molecule |
| chr16_+_12059050 | 5.04 |
ENST00000396495.3 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr19_+_50919056 | 5.02 |
ENST00000599632.1 |
CTD-2545M3.6 |
CTD-2545M3.6 |
| chr3_+_121774202 | 4.97 |
ENST00000469710.1 ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86 |
CD86 molecule |
| chr12_+_54891495 | 4.85 |
ENST00000293373.6 |
NCKAP1L |
NCK-associated protein 1-like |
| chr11_-_33891362 | 4.55 |
ENST00000395833.3 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr6_+_31582961 | 4.52 |
ENST00000376059.3 ENST00000337917.7 |
AIF1 |
allograft inflammatory factor 1 |
| chr12_+_7060432 | 4.29 |
ENST00000318974.9 ENST00000456013.1 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr17_+_65373531 | 4.23 |
ENST00000580974.1 |
PITPNC1 |
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr20_+_44746885 | 4.10 |
ENST00000372285.3 |
CD40 |
CD40 molecule, TNF receptor superfamily member 5 |
| chr20_+_44746939 | 4.07 |
ENST00000372276.3 |
CD40 |
CD40 molecule, TNF receptor superfamily member 5 |
| chr8_-_95449155 | 3.96 |
ENST00000481490.2 |
FSBP |
fibrinogen silencer binding protein |
| chr16_+_12059091 | 3.88 |
ENST00000562385.1 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr4_+_68424434 | 3.83 |
ENST00000265404.2 ENST00000396225.1 |
STAP1 |
signal transducing adaptor family member 1 |
| chr7_+_50344289 | 3.73 |
ENST00000413698.1 ENST00000359197.5 ENST00000331340.3 ENST00000357364.4 ENST00000343574.5 ENST00000349824.4 ENST00000346667.4 ENST00000440768.2 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
| chr10_+_11207438 | 3.68 |
ENST00000609692.1 ENST00000354897.3 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr1_+_161551101 | 3.60 |
ENST00000367962.4 ENST00000367960.5 ENST00000403078.3 ENST00000428605.2 |
FCGR2B |
Fc fragment of IgG, low affinity IIb, receptor (CD32) |
| chr17_+_65374075 | 3.59 |
ENST00000581322.1 |
PITPNC1 |
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr14_+_88471468 | 3.53 |
ENST00000267549.3 |
GPR65 |
G protein-coupled receptor 65 |
| chr9_+_93564191 | 3.52 |
ENST00000375747.1 |
SYK |
spleen tyrosine kinase |
| chr19_+_49838653 | 3.48 |
ENST00000598095.1 ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37 |
CD37 molecule |
| chr2_+_64751433 | 3.48 |
ENST00000238856.4 ENST00000422803.1 ENST00000238855.7 |
AFTPH |
aftiphilin |
| chr9_+_93564039 | 3.44 |
ENST00000375754.4 ENST00000375751.4 |
SYK |
spleen tyrosine kinase |
| chr20_+_30640004 | 3.43 |
ENST00000520553.1 ENST00000518730.1 ENST00000375852.2 |
HCK |
hemopoietic cell kinase |
| chr14_-_23288930 | 3.39 |
ENST00000554517.1 ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr19_-_13213662 | 3.31 |
ENST00000264824.4 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
| chr1_+_161475208 | 3.24 |
ENST00000367972.4 ENST00000271450.6 |
FCGR2A |
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
| chr5_+_112312416 | 3.22 |
ENST00000389063.2 |
DCP2 |
decapping mRNA 2 |
| chr17_-_73389737 | 3.18 |
ENST00000392563.1 |
GRB2 |
growth factor receptor-bound protein 2 |
| chr2_-_231084617 | 3.18 |
ENST00000409815.2 |
SP110 |
SP110 nuclear body protein |
| chr1_+_236305826 | 3.17 |
ENST00000366592.3 ENST00000366591.4 |
GPR137B |
G protein-coupled receptor 137B |
| chr17_-_73389854 | 3.16 |
ENST00000578961.1 ENST00000392564.1 ENST00000582582.1 |
GRB2 |
growth factor receptor-bound protein 2 |
| chr2_-_231084820 | 3.15 |
ENST00000258382.5 ENST00000338556.3 |
SP110 |
SP110 nuclear body protein |
| chr19_-_10446449 | 3.11 |
ENST00000592439.1 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr2_+_102972363 | 2.99 |
ENST00000409599.1 |
IL18R1 |
interleukin 18 receptor 1 |
| chr12_+_8276224 | 2.98 |
ENST00000229332.5 |
CLEC4A |
C-type lectin domain family 4, member A |
| chr8_-_77912431 | 2.95 |
ENST00000357039.4 ENST00000522527.1 |
PEX2 |
peroxisomal biogenesis factor 2 |
| chr18_-_52989525 | 2.88 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr2_-_231084659 | 2.87 |
ENST00000258381.6 ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110 |
SP110 nuclear body protein |
| chr7_-_37488834 | 2.87 |
ENST00000310758.4 |
ELMO1 |
engulfment and cell motility 1 |
| chr12_+_8276433 | 2.84 |
ENST00000345999.3 ENST00000352620.3 ENST00000360500.3 |
CLEC4A |
C-type lectin domain family 4, member A |
| chr1_+_14075865 | 2.83 |
ENST00000413440.1 |
PRDM2 |
PR domain containing 2, with ZNF domain |
| chrX_-_124097620 | 2.82 |
ENST00000371130.3 ENST00000422452.2 |
TENM1 |
teneurin transmembrane protein 1 |
| chr20_-_62710832 | 2.69 |
ENST00000395042.1 |
RGS19 |
regulator of G-protein signaling 19 |
| chr9_+_93589734 | 2.69 |
ENST00000375746.1 |
SYK |
spleen tyrosine kinase |
| chr10_+_11206925 | 2.67 |
ENST00000354440.2 ENST00000315874.4 ENST00000427450.1 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr11_-_3859089 | 2.59 |
ENST00000396979.1 |
RHOG |
ras homolog family member G |
| chr2_-_74405929 | 2.57 |
ENST00000396049.4 |
MOB1A |
MOB kinase activator 1A |
| chr1_-_159893507 | 2.54 |
ENST00000368096.1 |
TAGLN2 |
transgelin 2 |
| chrX_+_24072833 | 2.50 |
ENST00000253039.4 |
EIF2S3 |
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr8_+_98656693 | 2.48 |
ENST00000519934.1 |
MTDH |
metadherin |
| chr5_+_102455853 | 2.45 |
ENST00000515845.1 ENST00000321521.9 ENST00000507921.1 |
PPIP5K2 |
diphosphoinositol pentakisphosphate kinase 2 |
| chr14_+_102228123 | 2.43 |
ENST00000422945.2 ENST00000554442.1 ENST00000556260.2 ENST00000328724.5 ENST00000557268.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chr1_-_17380630 | 2.41 |
ENST00000375499.3 |
SDHB |
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr3_+_4535155 | 2.40 |
ENST00000544951.1 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr19_+_48828788 | 2.38 |
ENST00000594198.1 ENST00000597279.1 ENST00000593437.1 |
EMP3 |
epithelial membrane protein 3 |
| chr18_-_52989217 | 2.37 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr12_+_8276495 | 2.37 |
ENST00000546339.1 |
CLEC4A |
C-type lectin domain family 4, member A |
| chr3_-_16555150 | 2.35 |
ENST00000334133.4 |
RFTN1 |
raftlin, lipid raft linker 1 |
| chr1_+_14075903 | 2.31 |
ENST00000343137.4 ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2 |
PR domain containing 2, with ZNF domain |
| chrX_-_74376108 | 2.29 |
ENST00000339447.4 ENST00000373394.3 ENST00000529949.1 ENST00000534524.1 ENST00000253577.3 |
ABCB7 |
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
| chr21_-_46340884 | 2.26 |
ENST00000302347.5 ENST00000517819.1 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr6_-_133079022 | 2.26 |
ENST00000525289.1 ENST00000326499.6 |
VNN2 |
vanin 2 |
| chrX_-_30595959 | 2.23 |
ENST00000378962.3 |
CXorf21 |
chromosome X open reading frame 21 |
| chr10_+_28822636 | 2.17 |
ENST00000442148.1 ENST00000448193.1 |
WAC |
WW domain containing adaptor with coiled-coil |
| chr6_+_26251835 | 2.13 |
ENST00000356350.2 |
HIST1H2BH |
histone cluster 1, H2bh |
| chr11_-_60719213 | 2.11 |
ENST00000227880.3 |
SLC15A3 |
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr14_+_102276192 | 2.09 |
ENST00000557714.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chr21_-_46340770 | 2.09 |
ENST00000397854.3 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr5_+_102455968 | 2.09 |
ENST00000358359.3 |
PPIP5K2 |
diphosphoinositol pentakisphosphate kinase 2 |
| chr16_+_19183671 | 2.06 |
ENST00000562711.2 |
SYT17 |
synaptotagmin XVII |
| chr11_+_22696314 | 2.03 |
ENST00000532398.1 ENST00000433790.1 |
GAS2 |
growth arrest-specific 2 |
| chr3_-_176914238 | 2.01 |
ENST00000430069.1 ENST00000428970.1 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
| chrX_-_47489244 | 1.96 |
ENST00000469388.1 ENST00000396992.3 ENST00000377005.2 |
CFP |
complement factor properdin |
| chr19_+_48828582 | 1.96 |
ENST00000270221.6 ENST00000596315.1 |
EMP3 |
epithelial membrane protein 3 |
| chr5_-_142783175 | 1.95 |
ENST00000231509.3 ENST00000394464.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr20_-_62711259 | 1.93 |
ENST00000332298.5 |
RGS19 |
regulator of G-protein signaling 19 |
| chr9_+_127624387 | 1.86 |
ENST00000353214.2 |
ARPC5L |
actin related protein 2/3 complex, subunit 5-like |
| chr6_-_24936170 | 1.86 |
ENST00000538035.1 |
FAM65B |
family with sequence similarity 65, member B |
| chr22_-_32026810 | 1.85 |
ENST00000266095.5 ENST00000397500.1 |
PISD |
phosphatidylserine decarboxylase |
| chr3_+_23847394 | 1.85 |
ENST00000306627.3 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
| chrX_+_109246285 | 1.83 |
ENST00000372073.1 ENST00000372068.2 ENST00000288381.4 |
TMEM164 |
transmembrane protein 164 |
| chr21_+_34638656 | 1.79 |
ENST00000290200.2 |
IL10RB |
interleukin 10 receptor, beta |
| chr14_+_102276132 | 1.78 |
ENST00000350249.3 ENST00000557621.1 ENST00000556946.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chr21_-_34185944 | 1.72 |
ENST00000479548.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr3_+_23847432 | 1.69 |
ENST00000346855.3 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
| chr11_-_64889649 | 1.59 |
ENST00000434372.2 |
FAU |
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr11_+_65408273 | 1.56 |
ENST00000394227.3 |
SIPA1 |
signal-induced proliferation-associated 1 |
| chr2_+_113931513 | 1.54 |
ENST00000245796.6 ENST00000441564.3 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
| chr1_+_46713404 | 1.46 |
ENST00000371975.4 ENST00000469835.1 |
RAD54L |
RAD54-like (S. cerevisiae) |
| chr11_-_64512469 | 1.46 |
ENST00000377485.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr1_+_46713357 | 1.45 |
ENST00000442598.1 |
RAD54L |
RAD54-like (S. cerevisiae) |
| chr21_-_34186006 | 1.40 |
ENST00000490358.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr8_-_131028869 | 1.40 |
ENST00000518283.1 ENST00000519110.1 |
FAM49B |
family with sequence similarity 49, member B |
| chrX_+_11776701 | 1.40 |
ENST00000476743.1 ENST00000421368.2 ENST00000398527.2 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
| chrX_+_47420516 | 1.40 |
ENST00000377045.4 ENST00000290277.6 ENST00000377039.2 |
ARAF |
v-raf murine sarcoma 3611 viral oncogene homolog |
| chr8_+_74903580 | 1.38 |
ENST00000284818.2 ENST00000518893.1 |
LY96 |
lymphocyte antigen 96 |
| chr2_-_43019698 | 1.38 |
ENST00000431905.1 ENST00000294973.6 |
HAAO |
3-hydroxyanthranilate 3,4-dioxygenase |
| chr8_-_101962777 | 1.38 |
ENST00000395951.3 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr11_+_60048129 | 1.37 |
ENST00000355131.3 |
MS4A4A |
membrane-spanning 4-domains, subfamily A, member 4A |
| chr5_-_66492562 | 1.37 |
ENST00000256447.4 |
CD180 |
CD180 molecule |
| chr10_-_62149433 | 1.35 |
ENST00000280772.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr10_+_114133773 | 1.35 |
ENST00000354655.4 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
| chr5_-_142782862 | 1.32 |
ENST00000415690.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr7_+_139528952 | 1.32 |
ENST00000416849.2 ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
| chr15_-_34628951 | 1.32 |
ENST00000397707.2 ENST00000560611.1 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr1_-_231560790 | 1.30 |
ENST00000366641.3 |
EGLN1 |
egl-9 family hypoxia-inducible factor 1 |
| chr22_+_39410287 | 1.30 |
ENST00000381568.4 |
APOBEC3D |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D |
| chr9_-_127624194 | 1.29 |
ENST00000373570.4 ENST00000348462.3 |
RPL35 |
ribosomal protein L35 |
| chr19_+_3136115 | 1.27 |
ENST00000262958.3 |
GNA15 |
guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
| chr7_-_152133059 | 1.25 |
ENST00000262189.6 ENST00000355193.2 |
KMT2C |
lysine (K)-specific methyltransferase 2C |
| chr6_+_15246501 | 1.24 |
ENST00000341776.2 |
JARID2 |
jumonji, AT rich interactive domain 2 |
| chr1_+_44440575 | 1.22 |
ENST00000532642.1 ENST00000236067.4 ENST00000471859.2 |
ATP6V0B |
ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b |
| chrX_+_11777671 | 1.22 |
ENST00000380693.3 ENST00000380692.2 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
| chr6_+_31554779 | 1.21 |
ENST00000376090.2 |
LST1 |
leukocyte specific transcript 1 |
| chr2_-_153573965 | 1.21 |
ENST00000448428.1 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr17_-_76356148 | 1.21 |
ENST00000587578.1 ENST00000330871.2 |
SOCS3 |
suppressor of cytokine signaling 3 |
| chr3_-_160283348 | 1.20 |
ENST00000334256.4 |
KPNA4 |
karyopherin alpha 4 (importin alpha 3) |
| chr18_+_13218769 | 1.18 |
ENST00000399848.3 ENST00000361205.4 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
| chr4_+_113568207 | 1.17 |
ENST00000511529.1 |
LARP7 |
La ribonucleoprotein domain family, member 7 |
| chrX_-_129299847 | 1.16 |
ENST00000319908.3 ENST00000287295.3 |
AIFM1 |
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr1_+_32757668 | 1.16 |
ENST00000373548.3 |
HDAC1 |
histone deacetylase 1 |
| chr12_-_122018859 | 1.16 |
ENST00000536437.1 ENST00000377071.4 ENST00000538046.2 |
KDM2B |
lysine (K)-specific demethylase 2B |
| chr15_+_59279851 | 1.15 |
ENST00000348370.4 ENST00000434298.1 ENST00000559160.1 |
RNF111 |
ring finger protein 111 |
| chr17_+_7462103 | 1.15 |
ENST00000396545.4 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr14_+_102276209 | 1.15 |
ENST00000445439.3 ENST00000334743.5 ENST00000557095.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chr1_-_153919128 | 1.14 |
ENST00000361217.4 |
DENND4B |
DENN/MADD domain containing 4B |
| chr14_+_29234870 | 1.13 |
ENST00000382535.3 |
FOXG1 |
forkhead box G1 |
| chrX_+_11776278 | 1.13 |
ENST00000312196.4 ENST00000337339.2 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
| chr15_+_51973680 | 1.12 |
ENST00000542355.2 |
SCG3 |
secretogranin III |
| chr11_+_67250490 | 1.11 |
ENST00000528641.2 ENST00000279146.3 |
AIP |
aryl hydrocarbon receptor interacting protein |
| chr8_+_27950619 | 1.10 |
ENST00000542181.1 ENST00000524103.1 ENST00000537665.1 ENST00000380353.4 ENST00000520288.1 |
ELP3 |
elongator acetyltransferase complex subunit 3 |
| chr11_-_8680383 | 1.10 |
ENST00000299550.6 |
TRIM66 |
tripartite motif containing 66 |
| chr11_+_60048053 | 1.09 |
ENST00000337908.4 |
MS4A4A |
membrane-spanning 4-domains, subfamily A, member 4A |
| chr1_-_23886285 | 1.08 |
ENST00000374561.5 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr19_+_2476116 | 1.08 |
ENST00000215631.4 ENST00000587345.1 |
GADD45B |
growth arrest and DNA-damage-inducible, beta |
| chr7_+_23145884 | 1.05 |
ENST00000409689.1 ENST00000410047.1 |
KLHL7 |
kelch-like family member 7 |
| chr21_-_34185989 | 1.04 |
ENST00000487113.1 ENST00000382373.4 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr15_+_51973550 | 1.04 |
ENST00000220478.3 |
SCG3 |
secretogranin III |
| chr3_+_46395219 | 1.04 |
ENST00000445132.2 ENST00000292301.4 |
CCR2 |
chemokine (C-C motif) receptor 2 |
| chr2_+_173940442 | 1.03 |
ENST00000409176.2 ENST00000338983.3 ENST00000431503.2 |
MLTK |
Mitogen-activated protein kinase kinase kinase MLT |
| chr1_-_220445757 | 1.02 |
ENST00000358951.2 |
RAB3GAP2 |
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr1_+_166808692 | 1.00 |
ENST00000367876.4 |
POGK |
pogo transposable element with KRAB domain |
| chr11_-_64889529 | 1.00 |
ENST00000531743.1 ENST00000527548.1 ENST00000526555.1 ENST00000279259.3 |
FAU |
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr9_-_35618364 | 1.00 |
ENST00000378431.1 ENST00000378430.3 ENST00000259633.4 |
CD72 |
CD72 molecule |
| chr8_-_6420565 | 0.99 |
ENST00000338312.6 |
ANGPT2 |
angiopoietin 2 |
| chr5_+_177557997 | 0.98 |
ENST00000313386.4 ENST00000515098.1 ENST00000542098.1 ENST00000502814.1 ENST00000507457.1 ENST00000508647.1 |
RMND5B |
required for meiotic nuclear division 5 homolog B (S. cerevisiae) |
| chr11_+_64889773 | 0.98 |
ENST00000534078.1 ENST00000526171.1 ENST00000279242.2 ENST00000531705.1 ENST00000533943.1 |
MRPL49 |
mitochondrial ribosomal protein L49 |
| chr10_+_28822417 | 0.98 |
ENST00000428935.1 ENST00000420266.1 |
WAC |
WW domain containing adaptor with coiled-coil |
| chr11_-_102401469 | 0.97 |
ENST00000260227.4 |
MMP7 |
matrix metallopeptidase 7 (matrilysin, uterine) |
| chr8_-_6420759 | 0.97 |
ENST00000523120.1 |
ANGPT2 |
angiopoietin 2 |
| chr5_-_138210977 | 0.97 |
ENST00000274711.6 ENST00000521094.2 |
LRRTM2 |
leucine rich repeat transmembrane neuronal 2 |
| chr22_-_37880543 | 0.96 |
ENST00000442496.1 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr18_-_60986962 | 0.95 |
ENST00000333681.4 |
BCL2 |
B-cell CLL/lymphoma 2 |
| chr12_+_120105558 | 0.94 |
ENST00000229328.5 ENST00000541640.1 |
PRKAB1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr18_+_18943554 | 0.93 |
ENST00000580732.2 |
GREB1L |
growth regulation by estrogen in breast cancer-like |
| chr15_+_43885252 | 0.92 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr1_+_50575292 | 0.92 |
ENST00000371821.1 ENST00000371819.1 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr20_-_35274548 | 0.91 |
ENST00000262866.4 |
SLA2 |
Src-like-adaptor 2 |
| chr17_+_7461580 | 0.91 |
ENST00000483039.1 ENST00000396542.1 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr12_+_54447637 | 0.91 |
ENST00000609810.1 ENST00000430889.2 |
HOXC4 HOXC4 |
homeobox C4 Homeobox protein Hox-C4 |
| chr6_+_32146131 | 0.90 |
ENST00000375094.3 |
RNF5 |
ring finger protein 5, E3 ubiquitin protein ligase |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 41.9 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.5 | 24.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.4 | 32.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.4 | 6.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.4 | 5.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.3 | 22.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.3 | 16.7 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.2 | 9.0 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.2 | 10.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 9.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 4.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 2.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 2.4 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 3.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 3.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 6.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 5.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 3.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 12.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 5.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 2.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 2.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.1 | 32.4 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 5.3 | 15.8 | GO:0045404 | interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 3.4 | 10.1 | GO:0002605 | follicular B cell differentiation(GO:0002316) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) regulation of cytotoxic T cell degranulation(GO:0043317) negative regulation of cytotoxic T cell degranulation(GO:0043318) |
| 2.4 | 14.5 | GO:0045588 | positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 2.2 | 8.9 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 2.1 | 6.4 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 1.7 | 5.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 1.3 | 3.8 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 1.2 | 7.4 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 1.2 | 6.0 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 1.2 | 8.2 | GO:0033590 | response to cobalamin(GO:0033590) |
| 1.1 | 3.4 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 1.1 | 4.3 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 1.0 | 11.0 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 1.0 | 7.8 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.9 | 4.5 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.9 | 5.3 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.8 | 3.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.8 | 7.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.7 | 13.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.7 | 2.7 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.6 | 9.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.6 | 12.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.6 | 5.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.6 | 3.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.5 | 2.7 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.5 | 1.0 | GO:0002434 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) |
| 0.5 | 2.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.5 | 3.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.4 | 3.5 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.4 | 7.5 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.4 | 4.1 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.4 | 3.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.4 | 2.4 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.4 | 5.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.4 | 8.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.4 | 1.2 | GO:1904044 | response to aldosterone(GO:1904044) |
| 0.4 | 1.2 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.4 | 1.2 | GO:0021592 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 0.4 | 6.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.4 | 3.0 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.3 | 5.2 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.3 | 1.0 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.3 | 1.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.3 | 1.4 | GO:0072660 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 | 2.0 | GO:0060613 | fat pad development(GO:0060613) |
| 0.3 | 2.3 | GO:0032596 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.3 | 3.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.3 | 1.6 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.3 | 2.8 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.3 | 0.9 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.3 | 1.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.3 | 4.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 14.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 0.9 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.2 | 1.4 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.2 | 1.2 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.2 | 2.3 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.2 | 27.9 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.2 | 0.9 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 2.0 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.2 | 1.3 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.2 | 2.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 4.6 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.2 | 6.0 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.2 | 0.8 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 1.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 | 0.4 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.2 | 2.4 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.2 | 0.7 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.2 | 0.4 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.2 | 0.7 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.2 | 6.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 3.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.2 | 3.0 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 1.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 8.5 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.2 | 5.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 4.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.7 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 1.1 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.1 | 1.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 18.3 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 0.5 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 1.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.5 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) positive regulation of interferon-alpha biosynthetic process(GO:0045356) negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.1 | 5.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 2.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 3.5 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 7.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 11.4 | GO:0002819 | regulation of adaptive immune response(GO:0002819) |
| 0.1 | 0.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 1.2 | GO:0010990 | regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 11.6 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.1 | 0.4 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.1 | 0.4 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.1 | 0.8 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.1 | 1.0 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 1.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.1 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 15.6 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 1.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.1 | 0.9 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.1 | 1.1 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 1.1 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.1 | 2.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.2 | GO:2000611 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid hormone generation(GO:2000609) positive regulation of thyroid hormone generation(GO:2000611) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 1.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.2 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.1 | 8.5 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.1 | 0.4 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.7 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 1.2 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 1.3 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 1.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 5.2 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 1.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.8 | GO:0019054 | modulation by virus of host process(GO:0019054) |
| 0.0 | 7.4 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.9 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 1.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.9 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 2.4 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 3.6 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 1.7 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.3 | GO:0044849 | estrous cycle(GO:0044849) |
| 0.0 | 1.0 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.8 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.6 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0097205 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.0 | 0.7 | GO:0072431 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) |
| 0.0 | 0.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.3 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.5 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.1 | GO:0045008 | depyrimidination(GO:0045008) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 45.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.6 | 12.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.6 | 10.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.5 | 4.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.5 | 7.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.4 | 6.2 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.4 | 12.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.3 | 10.6 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.3 | 24.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.3 | 6.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.2 | 22.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.2 | 6.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 37.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 3.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 3.5 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 5.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 3.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.0 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 5.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 7.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 7.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 0.8 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 1.9 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 3.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 0.9 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.1 | 0.6 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.1 | 1.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.7 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 3.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 2.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.0 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 4.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 1.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 2.2 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.0 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 3.6 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.5 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 32.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 2.5 | 10.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 2.1 | 6.4 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 1.7 | 5.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 1.3 | 24.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 1.1 | 6.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.9 | 4.5 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.9 | 7.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.9 | 6.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.8 | 2.4 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.7 | 2.2 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.7 | 2.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.7 | 3.3 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.6 | 4.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.6 | 1.9 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.6 | 12.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.6 | 2.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.6 | 7.8 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.6 | 3.5 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.6 | 9.7 | GO:0035325 | receptor signaling protein tyrosine kinase activity(GO:0004716) Toll-like receptor binding(GO:0035325) |
| 0.6 | 8.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.5 | 5.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.5 | 2.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.4 | 13.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 1.3 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.4 | 5.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.4 | 7.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.4 | 1.2 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.4 | 3.2 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.3 | 1.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 7.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.3 | 14.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.3 | 0.9 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.3 | 3.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.3 | 2.9 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.3 | 8.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.3 | 1.8 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.2 | 2.7 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 1.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 1.4 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.2 | 2.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.2 | 10.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 12.2 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.2 | 2.7 | GO:0019864 | IgG binding(GO:0019864) |
| 0.2 | 3.0 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.2 | 4.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 0.7 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.2 | 5.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 6.6 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 6.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.8 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 2.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 3.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.4 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.1 | 17.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.4 | GO:0001016 | transcription factor activity, RNA polymerase III transcription factor binding(GO:0001007) RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.1 | 1.6 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 0.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.8 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 1.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 3.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.9 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 0.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 4.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.8 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 1.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 4.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 2.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 1.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 1.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 6.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.1 | 4.9 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.1 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 4.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.3 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.9 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 1.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 15.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 2.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 1.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.0 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.3 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 1.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.9 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 2.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 1.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.6 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 2.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 4.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 5.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.7 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 2.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 20.0 | GO:0004872 | receptor activity(GO:0004872) molecular transducer activity(GO:0060089) |
| 0.0 | 2.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 16.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 1.2 | 8.2 | GO:0043196 | varicosity(GO:0043196) |
| 1.0 | 2.9 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.9 | 5.7 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.9 | 4.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.8 | 6.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.7 | 13.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.7 | 4.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.6 | 2.4 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.5 | 3.5 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.4 | 5.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.4 | 5.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.4 | 4.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.4 | 2.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.3 | 4.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 1.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.3 | 3.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.3 | 3.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.3 | 1.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 2.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 0.9 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.2 | 3.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.2 | 3.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 6.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 1.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 11.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 11.9 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 1.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 2.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.8 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 1.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 5.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.5 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 10.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 17.5 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 1.4 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.9 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.7 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.9 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 9.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 7.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 4.8 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.1 | 5.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 1.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 2.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 2.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 6.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 1.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 6.9 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.0 | 12.7 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 17.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.5 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.9 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 2.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 5.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 3.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 1.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 3.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 1.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 3.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.5 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 34.5 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 1.4 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 6.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 3.3 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 2.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 1.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 1.7 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.7 | GO:0005884 | actin filament(GO:0005884) |