Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
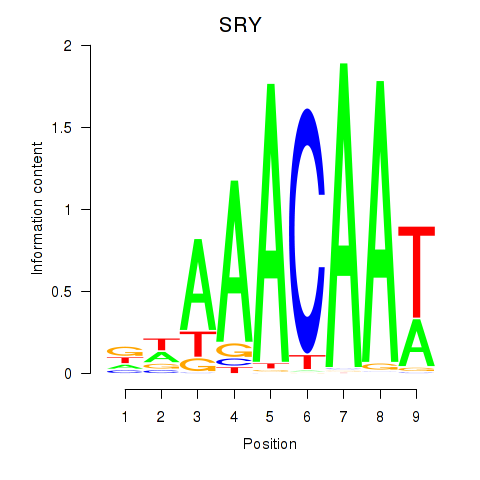
Results for SRY
Z-value: 1.11
Transcription factors associated with SRY
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SRY
|
ENSG00000184895.6 | SRY |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SRY | hg19_v2_chrY_-_2655644_2655740 | -0.58 | 1.9e-02 | Click! |
Activity profile of SRY motif
Sorted Z-values of SRY motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SRY
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_11206925 | 4.22 |
ENST00000354440.2 ENST00000315874.4 ENST00000427450.1 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr10_+_70847852 | 4.13 |
ENST00000242465.3 |
SRGN |
serglycin |
| chr13_-_46716969 | 2.47 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr2_-_152146385 | 2.08 |
ENST00000414946.1 ENST00000243346.5 |
NMI |
N-myc (and STAT) interactor |
| chr3_-_141868293 | 1.91 |
ENST00000317104.7 ENST00000494358.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr3_-_141868357 | 1.70 |
ENST00000489671.1 ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr10_+_11207438 | 1.66 |
ENST00000609692.1 ENST00000354897.3 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr2_-_192711968 | 1.59 |
ENST00000304141.4 |
SDPR |
serum deprivation response |
| chr4_-_103266626 | 1.58 |
ENST00000356736.4 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr4_-_105416039 | 1.54 |
ENST00000394767.2 |
CXXC4 |
CXXC finger protein 4 |
| chr7_-_99698338 | 1.49 |
ENST00000354230.3 ENST00000425308.1 |
MCM7 |
minichromosome maintenance complex component 7 |
| chr18_-_53303123 | 1.43 |
ENST00000569357.1 ENST00000565124.1 ENST00000398339.1 |
TCF4 |
transcription factor 4 |
| chr4_-_73434498 | 1.37 |
ENST00000286657.4 |
ADAMTS3 |
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr4_-_109090106 | 1.37 |
ENST00000379951.2 |
LEF1 |
lymphoid enhancer-binding factor 1 |
| chr11_-_10830463 | 1.34 |
ENST00000527419.1 ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr6_-_32557610 | 1.33 |
ENST00000360004.5 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
| chr18_-_53257027 | 1.29 |
ENST00000568740.1 ENST00000564403.2 ENST00000537578.1 |
TCF4 |
transcription factor 4 |
| chr1_+_33116765 | 1.25 |
ENST00000544435.1 ENST00000373485.1 ENST00000458695.2 ENST00000490500.1 ENST00000445722.2 |
RBBP4 |
retinoblastoma binding protein 4 |
| chr11_-_10829851 | 1.18 |
ENST00000532082.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr17_-_56082455 | 1.18 |
ENST00000578794.1 |
RP11-159D12.5 |
Uncharacterized protein |
| chr5_-_137674000 | 1.17 |
ENST00000510119.1 ENST00000513970.1 |
CDC25C |
cell division cycle 25C |
| chr4_-_109089573 | 1.10 |
ENST00000265165.1 |
LEF1 |
lymphoid enhancer-binding factor 1 |
| chr3_+_69812701 | 1.10 |
ENST00000472437.1 |
MITF |
microphthalmia-associated transcription factor |
| chr22_+_19710468 | 1.09 |
ENST00000366425.3 |
GP1BB |
glycoprotein Ib (platelet), beta polypeptide |
| chr12_-_102591604 | 1.09 |
ENST00000329406.4 |
PMCH |
pro-melanin-concentrating hormone |
| chr15_-_55562479 | 1.07 |
ENST00000564609.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr12_-_102872317 | 1.01 |
ENST00000424202.2 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr15_-_55563072 | 0.98 |
ENST00000567380.1 ENST00000565972.1 ENST00000569493.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr8_+_21777159 | 0.90 |
ENST00000434536.1 ENST00000252512.9 |
XPO7 |
exportin 7 |
| chr21_-_19191703 | 0.88 |
ENST00000284881.4 ENST00000400559.3 ENST00000400558.3 |
C21orf91 |
chromosome 21 open reading frame 91 |
| chr12_-_102874416 | 0.86 |
ENST00000392904.1 ENST00000337514.6 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr17_-_40428359 | 0.85 |
ENST00000293328.3 |
STAT5B |
signal transducer and activator of transcription 5B |
| chr22_+_40297079 | 0.85 |
ENST00000344138.4 ENST00000543252.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr11_+_112832090 | 0.85 |
ENST00000533760.1 |
NCAM1 |
neural cell adhesion molecule 1 |
| chr9_+_139874683 | 0.84 |
ENST00000444903.1 |
PTGDS |
prostaglandin D2 synthase 21kDa (brain) |
| chr10_-_62149433 | 0.83 |
ENST00000280772.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr9_-_128246769 | 0.83 |
ENST00000444226.1 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
| chr2_+_191273052 | 0.83 |
ENST00000417958.1 ENST00000432036.1 ENST00000392328.1 |
MFSD6 |
major facilitator superfamily domain containing 6 |
| chr18_-_52969844 | 0.83 |
ENST00000561831.3 |
TCF4 |
transcription factor 4 |
| chr13_+_102142296 | 0.82 |
ENST00000376162.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr2_-_157198860 | 0.82 |
ENST00000409572.1 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
| chr3_+_133118839 | 0.82 |
ENST00000302334.2 |
BFSP2 |
beaded filament structural protein 2, phakinin |
| chr22_+_40297105 | 0.79 |
ENST00000540310.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr12_-_102874378 | 0.79 |
ENST00000456098.1 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr17_-_46703826 | 0.76 |
ENST00000550387.1 ENST00000311177.5 |
HOXB9 |
homeobox B9 |
| chr2_-_190927447 | 0.74 |
ENST00000260950.4 |
MSTN |
myostatin |
| chr1_+_100316041 | 0.73 |
ENST00000370165.3 ENST00000370163.3 ENST00000294724.4 |
AGL |
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr15_-_55562582 | 0.70 |
ENST00000396307.2 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr18_-_52989217 | 0.70 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr11_+_112832202 | 0.65 |
ENST00000534015.1 |
NCAM1 |
neural cell adhesion molecule 1 |
| chr13_-_99910673 | 0.64 |
ENST00000397473.2 ENST00000397470.2 |
GPR18 |
G protein-coupled receptor 18 |
| chr4_+_170581213 | 0.63 |
ENST00000507875.1 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr12_-_92539614 | 0.61 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr10_-_61900762 | 0.61 |
ENST00000355288.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr8_-_95449155 | 0.60 |
ENST00000481490.2 |
FSBP |
fibrinogen silencer binding protein |
| chr12_-_8815404 | 0.59 |
ENST00000359478.2 ENST00000396549.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr18_-_52989525 | 0.59 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr4_-_76598296 | 0.58 |
ENST00000395719.3 |
G3BP2 |
GTPase activating protein (SH3 domain) binding protein 2 |
| chr2_-_37899323 | 0.58 |
ENST00000295324.3 ENST00000457889.1 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
| chr3_+_178866199 | 0.57 |
ENST00000263967.3 |
PIK3CA |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr14_-_23904861 | 0.56 |
ENST00000355349.3 |
MYH7 |
myosin, heavy chain 7, cardiac muscle, beta |
| chr4_-_76598544 | 0.56 |
ENST00000515457.1 ENST00000357854.3 |
G3BP2 |
GTPase activating protein (SH3 domain) binding protein 2 |
| chr1_+_165797024 | 0.54 |
ENST00000372212.4 |
UCK2 |
uridine-cytidine kinase 2 |
| chr12_-_102874102 | 0.53 |
ENST00000392905.2 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr6_+_122720681 | 0.53 |
ENST00000368455.4 ENST00000452194.1 |
HSF2 |
heat shock transcription factor 2 |
| chr4_+_41614909 | 0.52 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr4_-_83280767 | 0.51 |
ENST00000514671.1 |
HNRNPD |
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr5_-_146435694 | 0.51 |
ENST00000356826.3 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
| chr1_-_40367668 | 0.49 |
ENST00000397332.2 ENST00000429311.1 |
MYCL |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr11_+_112832133 | 0.47 |
ENST00000524665.1 |
NCAM1 |
neural cell adhesion molecule 1 |
| chr16_+_6069072 | 0.47 |
ENST00000547605.1 ENST00000550418.1 ENST00000553186.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr8_+_70404996 | 0.46 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr17_-_10452929 | 0.46 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr3_-_33700589 | 0.46 |
ENST00000461133.3 ENST00000496954.2 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr14_-_57960545 | 0.46 |
ENST00000526336.1 ENST00000216445.3 |
C14orf105 |
chromosome 14 open reading frame 105 |
| chr3_+_178253993 | 0.46 |
ENST00000420517.2 ENST00000452583.1 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr13_-_74708372 | 0.45 |
ENST00000377666.4 |
KLF12 |
Kruppel-like factor 12 |
| chr12_-_102874330 | 0.44 |
ENST00000307046.8 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr10_-_62332357 | 0.44 |
ENST00000503366.1 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr4_-_164534657 | 0.43 |
ENST00000339875.5 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr11_-_111637083 | 0.43 |
ENST00000427203.2 ENST00000341980.6 ENST00000311129.5 ENST00000393055.2 ENST00000426998.2 ENST00000527614.1 |
PPP2R1B |
protein phosphatase 2, regulatory subunit A, beta |
| chr16_+_6069586 | 0.42 |
ENST00000547372.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr14_-_23288930 | 0.42 |
ENST00000554517.1 ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr1_+_147374915 | 0.42 |
ENST00000240986.4 |
GJA8 |
gap junction protein, alpha 8, 50kDa |
| chr9_-_124989804 | 0.41 |
ENST00000373755.2 ENST00000373754.2 |
LHX6 |
LIM homeobox 6 |
| chrX_+_78003204 | 0.41 |
ENST00000435339.3 ENST00000514744.1 |
LPAR4 |
lysophosphatidic acid receptor 4 |
| chr17_-_49124230 | 0.40 |
ENST00000510283.1 ENST00000510855.1 |
SPAG9 |
sperm associated antigen 9 |
| chr1_-_53018654 | 0.40 |
ENST00000257177.4 ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11 |
zinc finger, CCHC domain containing 11 |
| chr21_-_19775973 | 0.39 |
ENST00000284885.3 |
TMPRSS15 |
transmembrane protease, serine 15 |
| chr2_-_220042825 | 0.39 |
ENST00000409789.1 |
CNPPD1 |
cyclin Pas1/PHO80 domain containing 1 |
| chr12_-_8815215 | 0.38 |
ENST00000544889.1 ENST00000543369.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chrX_-_110655306 | 0.38 |
ENST00000371993.2 |
DCX |
doublecortin |
| chr7_+_106505912 | 0.38 |
ENST00000359195.3 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr1_+_50571949 | 0.37 |
ENST00000357083.4 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chrX_-_106959631 | 0.37 |
ENST00000486554.1 ENST00000372390.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr17_+_36861735 | 0.37 |
ENST00000378137.5 ENST00000325718.7 |
MLLT6 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr1_-_193155729 | 0.37 |
ENST00000367434.4 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr2_+_120517174 | 0.36 |
ENST00000263708.2 |
PTPN4 |
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr22_+_31795509 | 0.36 |
ENST00000331457.4 |
DRG1 |
developmentally regulated GTP binding protein 1 |
| chr7_+_28452130 | 0.36 |
ENST00000357727.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr4_+_71588372 | 0.33 |
ENST00000536664.1 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr2_+_208423891 | 0.33 |
ENST00000448277.1 ENST00000457101.1 |
CREB1 |
cAMP responsive element binding protein 1 |
| chr17_+_8924837 | 0.32 |
ENST00000173229.2 |
NTN1 |
netrin 1 |
| chr3_-_33700933 | 0.32 |
ENST00000480013.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr1_-_117210290 | 0.32 |
ENST00000369483.1 ENST00000369486.3 |
IGSF3 |
immunoglobulin superfamily, member 3 |
| chr7_+_129015484 | 0.32 |
ENST00000490911.1 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
| chrX_-_110655391 | 0.31 |
ENST00000356915.2 ENST00000356220.3 |
DCX |
doublecortin |
| chr14_-_51027838 | 0.31 |
ENST00000555216.1 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr12_+_94542459 | 0.31 |
ENST00000258526.4 |
PLXNC1 |
plexin C1 |
| chr6_-_161085291 | 0.31 |
ENST00000316300.5 |
LPA |
lipoprotein, Lp(a) |
| chr12_-_8815299 | 0.30 |
ENST00000535336.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr7_+_142985308 | 0.30 |
ENST00000310447.5 |
CASP2 |
caspase 2, apoptosis-related cysteine peptidase |
| chr12_+_31477250 | 0.30 |
ENST00000313737.4 |
AC024940.1 |
AC024940.1 |
| chr1_+_153232160 | 0.29 |
ENST00000368742.3 |
LOR |
loricrin |
| chr2_+_86668464 | 0.29 |
ENST00000409064.1 |
KDM3A |
lysine (K)-specific demethylase 3A |
| chr1_+_159409512 | 0.29 |
ENST00000423932.3 |
OR10J1 |
olfactory receptor, family 10, subfamily J, member 1 |
| chr12_-_8815477 | 0.28 |
ENST00000433590.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr14_+_58894706 | 0.28 |
ENST00000261244.5 |
KIAA0586 |
KIAA0586 |
| chr5_-_146461027 | 0.27 |
ENST00000394410.2 ENST00000508267.1 ENST00000504198.1 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
| chr10_-_99094458 | 0.27 |
ENST00000371019.2 |
FRAT2 |
frequently rearranged in advanced T-cell lymphomas 2 |
| chr15_+_49715293 | 0.27 |
ENST00000267843.4 ENST00000560270.1 |
FGF7 |
fibroblast growth factor 7 |
| chr2_+_162101247 | 0.26 |
ENST00000439050.1 ENST00000436506.1 |
AC009299.3 |
AC009299.3 |
| chr20_-_35492048 | 0.26 |
ENST00000237536.4 |
SOGA1 |
suppressor of glucose, autophagy associated 1 |
| chr14_-_58894332 | 0.26 |
ENST00000395159.2 |
TIMM9 |
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr1_-_115292591 | 0.26 |
ENST00000438362.2 |
CSDE1 |
cold shock domain containing E1, RNA-binding |
| chr1_+_235490659 | 0.25 |
ENST00000488594.1 |
GGPS1 |
geranylgeranyl diphosphate synthase 1 |
| chr3_-_57233966 | 0.25 |
ENST00000473921.1 ENST00000295934.3 |
HESX1 |
HESX homeobox 1 |
| chr14_-_61124977 | 0.25 |
ENST00000554986.1 |
SIX1 |
SIX homeobox 1 |
| chr9_-_23826298 | 0.25 |
ENST00000380117.1 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr2_-_2334888 | 0.24 |
ENST00000428368.2 ENST00000399161.2 |
MYT1L |
myelin transcription factor 1-like |
| chr8_+_24151620 | 0.24 |
ENST00000437154.2 |
ADAM28 |
ADAM metallopeptidase domain 28 |
| chr8_-_87755878 | 0.24 |
ENST00000320005.5 |
CNGB3 |
cyclic nucleotide gated channel beta 3 |
| chr9_+_77112244 | 0.24 |
ENST00000376896.3 |
RORB |
RAR-related orphan receptor B |
| chr15_+_59397275 | 0.24 |
ENST00000288207.2 |
CCNB2 |
cyclin B2 |
| chr5_-_146435572 | 0.23 |
ENST00000394414.1 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
| chr12_-_56352368 | 0.22 |
ENST00000549404.1 |
PMEL |
premelanosome protein |
| chr6_+_139094657 | 0.21 |
ENST00000332797.6 |
CCDC28A |
coiled-coil domain containing 28A |
| chr1_+_50574585 | 0.21 |
ENST00000371824.1 ENST00000371823.4 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr17_+_27573875 | 0.21 |
ENST00000225387.3 |
CRYBA1 |
crystallin, beta A1 |
| chr1_-_156217822 | 0.20 |
ENST00000368270.1 |
PAQR6 |
progestin and adipoQ receptor family member VI |
| chr18_+_18943554 | 0.20 |
ENST00000580732.2 |
GREB1L |
growth regulation by estrogen in breast cancer-like |
| chr8_+_24151553 | 0.20 |
ENST00000265769.4 ENST00000540823.1 ENST00000397649.3 |
ADAM28 |
ADAM metallopeptidase domain 28 |
| chr8_+_57124245 | 0.20 |
ENST00000521831.1 ENST00000355315.3 ENST00000303759.3 ENST00000517636.1 ENST00000517933.1 ENST00000518801.1 ENST00000523975.1 ENST00000396723.5 ENST00000523061.1 ENST00000521524.1 |
CHCHD7 |
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr2_+_27070964 | 0.20 |
ENST00000288699.6 |
DPYSL5 |
dihydropyrimidinase-like 5 |
| chrX_+_15525426 | 0.20 |
ENST00000342014.6 |
BMX |
BMX non-receptor tyrosine kinase |
| chr1_-_115053781 | 0.19 |
ENST00000358465.2 ENST00000369543.2 |
TRIM33 |
tripartite motif containing 33 |
| chr6_+_126070726 | 0.19 |
ENST00000368364.3 |
HEY2 |
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr1_+_25664408 | 0.19 |
ENST00000374358.4 |
TMEM50A |
transmembrane protein 50A |
| chr5_-_131132658 | 0.19 |
ENST00000514667.1 ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3 FNIP1 |
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr1_-_156217829 | 0.19 |
ENST00000356983.2 ENST00000335852.1 ENST00000340183.5 ENST00000540423.1 |
PAQR6 |
progestin and adipoQ receptor family member VI |
| chr11_-_10879572 | 0.19 |
ENST00000413761.2 |
ZBED5 |
zinc finger, BED-type containing 5 |
| chr19_+_13134772 | 0.18 |
ENST00000587760.1 ENST00000585575.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr10_+_115312766 | 0.18 |
ENST00000351270.3 |
HABP2 |
hyaluronan binding protein 2 |
| chr1_-_156217875 | 0.18 |
ENST00000292291.5 |
PAQR6 |
progestin and adipoQ receptor family member VI |
| chr11_-_10879593 | 0.18 |
ENST00000528289.1 ENST00000432999.2 |
ZBED5 |
zinc finger, BED-type containing 5 |
| chrX_+_99839799 | 0.17 |
ENST00000373031.4 |
TNMD |
tenomodulin |
| chrX_+_123095546 | 0.17 |
ENST00000371157.3 ENST00000371145.3 ENST00000371144.3 |
STAG2 |
stromal antigen 2 |
| chr21_-_32931290 | 0.17 |
ENST00000286827.3 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
| chr16_-_4323015 | 0.17 |
ENST00000204517.6 |
TFAP4 |
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr9_+_27109133 | 0.17 |
ENST00000519097.1 ENST00000380036.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chr14_-_60097297 | 0.16 |
ENST00000395090.1 |
RTN1 |
reticulon 1 |
| chr5_-_59481406 | 0.16 |
ENST00000546160.1 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr11_+_128563652 | 0.16 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_-_116381918 | 0.16 |
ENST00000606080.1 |
FRK |
fyn-related kinase |
| chrX_+_28605516 | 0.16 |
ENST00000378993.1 |
IL1RAPL1 |
interleukin 1 receptor accessory protein-like 1 |
| chr6_-_79787902 | 0.15 |
ENST00000275034.4 |
PHIP |
pleckstrin homology domain interacting protein |
| chr11_+_128562372 | 0.15 |
ENST00000344954.6 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr1_+_215256467 | 0.15 |
ENST00000391894.2 ENST00000444842.2 |
KCNK2 |
potassium channel, subfamily K, member 2 |
| chr1_-_217250231 | 0.15 |
ENST00000493748.1 ENST00000463665.1 |
ESRRG |
estrogen-related receptor gamma |
| chr7_+_106505696 | 0.15 |
ENST00000440650.2 ENST00000496166.1 ENST00000473541.1 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr2_+_109204743 | 0.15 |
ENST00000332345.6 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr6_+_72596604 | 0.15 |
ENST00000348717.5 ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr6_-_152489484 | 0.15 |
ENST00000354674.4 ENST00000539504.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr9_-_37465396 | 0.14 |
ENST00000307750.4 |
ZBTB5 |
zinc finger and BTB domain containing 5 |
| chr11_-_30608413 | 0.14 |
ENST00000528686.1 |
MPPED2 |
metallophosphoesterase domain containing 2 |
| chr12_-_86650045 | 0.14 |
ENST00000604798.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr5_-_146435501 | 0.14 |
ENST00000336640.6 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
| chr11_+_67007518 | 0.13 |
ENST00000530342.1 ENST00000308783.5 |
KDM2A |
lysine (K)-specific demethylase 2A |
| chr2_+_10262857 | 0.13 |
ENST00000304567.5 |
RRM2 |
ribonucleotide reductase M2 |
| chr2_+_86947296 | 0.13 |
ENST00000283632.4 |
RMND5A |
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr2_-_219925189 | 0.13 |
ENST00000295731.6 |
IHH |
indian hedgehog |
| chr1_+_41174988 | 0.13 |
ENST00000372652.1 |
NFYC |
nuclear transcription factor Y, gamma |
| chr7_-_27219849 | 0.13 |
ENST00000396344.4 |
HOXA10 |
homeobox A10 |
| chr12_-_86650077 | 0.12 |
ENST00000552808.2 ENST00000547225.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr8_-_57123815 | 0.12 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr9_-_123476612 | 0.12 |
ENST00000426959.1 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr4_-_103746683 | 0.12 |
ENST00000504211.1 ENST00000508476.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr6_+_108882069 | 0.11 |
ENST00000406360.1 |
FOXO3 |
forkhead box O3 |
| chr11_+_22359562 | 0.11 |
ENST00000263160.3 |
SLC17A6 |
solute carrier family 17 (vesicular glutamate transporter), member 6 |
| chr11_-_82708435 | 0.11 |
ENST00000525117.1 ENST00000532548.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr1_-_43855444 | 0.10 |
ENST00000372455.4 |
MED8 |
mediator complex subunit 8 |
| chr14_+_58894103 | 0.10 |
ENST00000354386.6 ENST00000556134.1 |
KIAA0586 |
KIAA0586 |
| chr7_+_128399002 | 0.10 |
ENST00000493278.1 |
CALU |
calumenin |
| chr14_+_22985251 | 0.10 |
ENST00000390510.1 |
TRAJ27 |
T cell receptor alpha joining 27 |
| chr4_+_71263599 | 0.10 |
ENST00000399575.2 |
PROL1 |
proline rich, lacrimal 1 |
| chr1_+_164529004 | 0.10 |
ENST00000559240.1 ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr19_-_46272106 | 0.10 |
ENST00000560168.1 |
SIX5 |
SIX homeobox 5 |
| chrX_+_84499038 | 0.10 |
ENST00000373165.3 |
ZNF711 |
zinc finger protein 711 |
| chr13_-_114843416 | 0.10 |
ENST00000389544.4 |
RASA3 |
RAS p21 protein activator 3 |
| chr12_-_122985494 | 0.10 |
ENST00000336229.4 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.6 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.3 | 1.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.2 | 2.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 2.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 4.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 0.5 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 0.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 2.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 2.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.6 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 1.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.3 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 1.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 4.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 3.7 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 2.2 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.4 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.8 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.8 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 1.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.2 | 2.5 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 5.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.4 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 2.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.4 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.7 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 3.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.3 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 4.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 1.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.3 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 1.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 2.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 2.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 2.0 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 1.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 2.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 4.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 1.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 2.0 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 4.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.7 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 1.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 4.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 8.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 0.8 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 5.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.8 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.7 | 3.6 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.7 | 2.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.6 | 2.5 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.5 | 1.9 | GO:0072660 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.5 | 2.7 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.3 | 0.8 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.3 | 1.3 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.3 | 1.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.2 | 1.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.2 | 0.8 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.2 | 0.6 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.2 | 0.7 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.2 | 0.2 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.2 | 0.5 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.2 | 1.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 5.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 2.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.6 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 0.8 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.5 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.6 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 1.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.7 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.8 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.3 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 0.3 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.5 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.1 | 0.3 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 0.2 | GO:0061055 | myotome development(GO:0061055) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 3.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.6 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.1 | 0.9 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.1 | 0.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.3 | GO:0034670 | chemotaxis to arachidonic acid(GO:0034670) response to arachidonic acid(GO:1904550) |
| 0.1 | 0.8 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.5 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 1.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 1.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.4 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0035234 | luteolysis(GO:0001554) ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.4 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 1.1 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.1 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.3 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.6 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 1.1 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 1.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 2.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0052026 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.3 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.0 | 0.7 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 4.9 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 1.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.3 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 2.0 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.2 | GO:0048753 | melanosome organization(GO:0032438) pigment granule organization(GO:0048753) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.3 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.0 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |