Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for STAT4
Z-value: 0.95
Transcription factors associated with STAT4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT4
|
ENSG00000138378.13 | STAT4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT4 | hg19_v2_chr2_-_192016316_192016325 | 0.05 | 8.6e-01 | Click! |
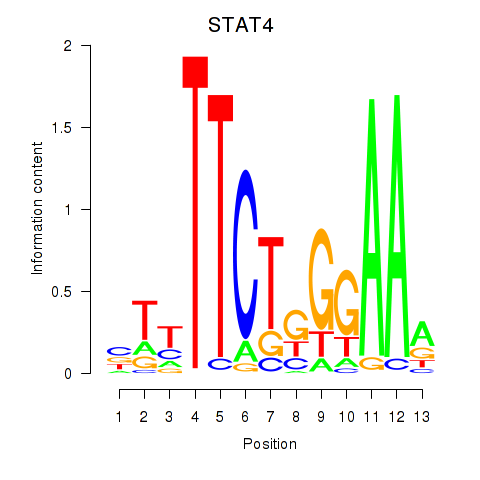
Activity profile of STAT4 motif
Sorted Z-values of STAT4 motif
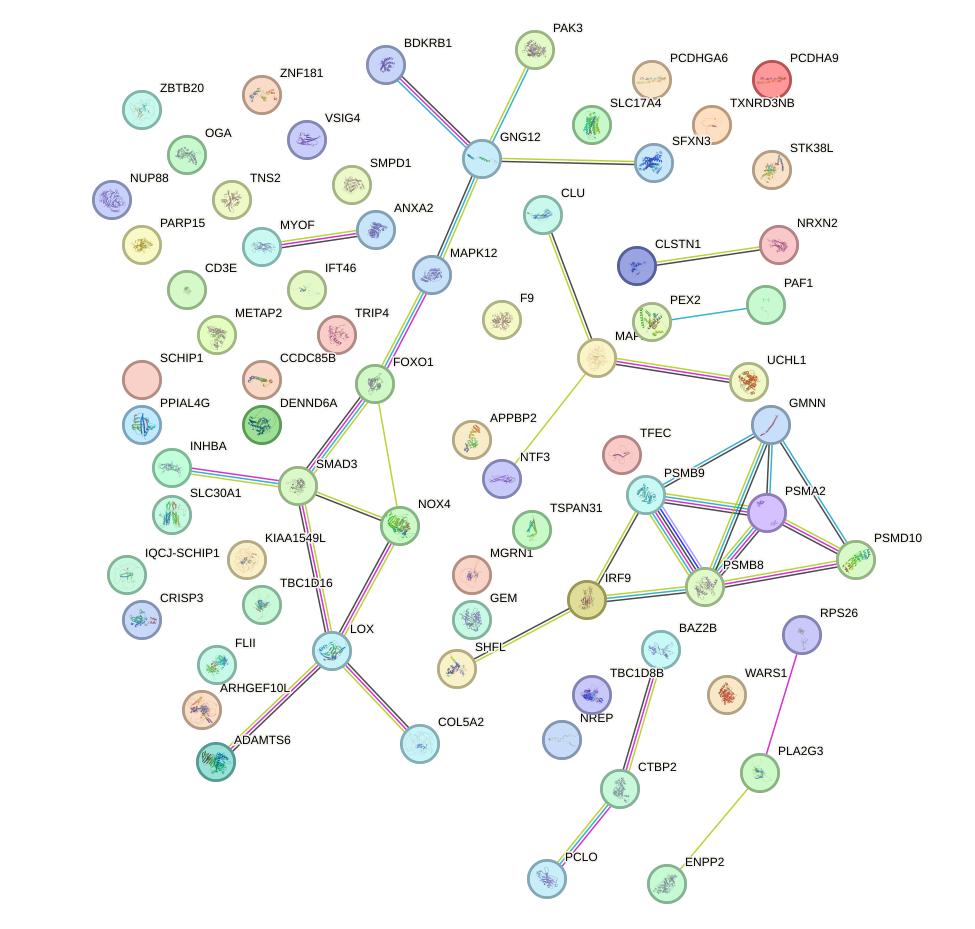
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_190044480 | 3.57 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr1_-_68299130 | 3.52 |
ENST00000370982.3 |
GNG12 |
guanine nucleotide binding protein (G protein), gamma 12 |
| chr10_-_95241951 | 3.17 |
ENST00000358334.5 ENST00000359263.4 ENST00000371488.3 |
MYOF |
myoferlin |
| chr10_-_95242044 | 2.94 |
ENST00000371501.4 ENST00000371502.4 ENST00000371489.1 |
MYOF |
myoferlin |
| chr4_+_41258786 | 2.42 |
ENST00000503431.1 ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr15_+_63340858 | 2.23 |
ENST00000560615.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr15_+_63340734 | 2.16 |
ENST00000560959.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr15_+_63340775 | 2.05 |
ENST00000559281.1 ENST00000317516.7 |
TPM1 |
tropomyosin 1 (alpha) |
| chr15_+_63340647 | 2.03 |
ENST00000404484.4 |
TPM1 |
tropomyosin 1 (alpha) |
| chr19_+_35168567 | 1.99 |
ENST00000457781.2 ENST00000505163.1 ENST00000505242.1 ENST00000423823.2 ENST00000507959.1 ENST00000446502.2 |
ZNF302 |
zinc finger protein 302 |
| chr10_-_126847276 | 1.96 |
ENST00000531469.1 |
CTBP2 |
C-terminal binding protein 2 |
| chr19_+_35168633 | 1.93 |
ENST00000505365.2 |
ZNF302 |
zinc finger protein 302 |
| chr6_+_36646435 | 1.92 |
ENST00000244741.5 ENST00000405375.1 ENST00000373711.2 |
CDKN1A |
cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| chr5_+_140753444 | 1.85 |
ENST00000517434.1 |
PCDHGA6 |
protocadherin gamma subfamily A, 6 |
| chr17_-_46682321 | 1.80 |
ENST00000225648.3 ENST00000484302.2 |
HOXB6 |
homeobox B6 |
| chr7_-_41740181 | 1.80 |
ENST00000442711.1 |
INHBA |
inhibin, beta A |
| chr19_+_35225060 | 1.76 |
ENST00000599244.1 ENST00000392232.3 |
ZNF181 |
zinc finger protein 181 |
| chr3_+_159570722 | 1.68 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr5_-_121413974 | 1.52 |
ENST00000231004.4 |
LOX |
lysyl oxidase |
| chr5_-_111093406 | 1.42 |
ENST00000379671.3 |
NREP |
neuronal regeneration related protein |
| chr5_-_111092873 | 1.38 |
ENST00000509025.1 ENST00000515855.1 |
NREP |
neuronal regeneration related protein |
| chr5_-_111093167 | 1.38 |
ENST00000446294.2 ENST00000419114.2 |
NREP |
neuronal regeneration related protein |
| chr5_-_111092930 | 1.37 |
ENST00000257435.7 |
NREP |
neuronal regeneration related protein |
| chr14_-_30396948 | 1.33 |
ENST00000331968.5 |
PRKD1 |
protein kinase D1 |
| chr15_-_60690932 | 1.05 |
ENST00000559818.1 |
ANXA2 |
annexin A2 |
| chr5_-_111093081 | 1.05 |
ENST00000453526.2 ENST00000509427.1 |
NREP |
neuronal regeneration related protein |
| chr10_+_102790980 | 1.04 |
ENST00000393459.1 ENST00000224807.5 |
SFXN3 |
sideroflexin 3 |
| chr3_-_114866084 | 1.03 |
ENST00000357258.3 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr8_-_120685608 | 1.02 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr5_-_111093340 | 1.00 |
ENST00000508870.1 |
NREP |
neuronal regeneration related protein |
| chr15_+_67458357 | 0.90 |
ENST00000537194.2 |
SMAD3 |
SMAD family member 3 |
| chr6_-_32811771 | 0.85 |
ENST00000395339.3 ENST00000374882.3 |
PSMB8 |
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr12_+_14518598 | 0.83 |
ENST00000261168.4 ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP |
activating transcription factor 7 interacting protein |
| chr19_+_44669221 | 0.83 |
ENST00000590578.1 ENST00000589160.1 ENST00000337433.5 ENST00000586286.1 ENST00000585560.1 ENST00000586914.1 ENST00000588883.1 ENST00000413984.2 ENST00000588742.1 ENST00000300823.6 ENST00000585678.1 ENST00000586203.1 ENST00000590467.1 ENST00000588795.1 ENST00000588127.1 |
ZNF226 |
zinc finger protein 226 |
| chr11_-_89224139 | 0.79 |
ENST00000413594.2 |
NOX4 |
NADPH oxidase 4 |
| chrX_+_54834159 | 0.74 |
ENST00000375053.2 ENST00000347546.4 ENST00000375062.4 |
MAGED2 |
melanoma antigen family D, 2 |
| chr14_+_24630465 | 0.74 |
ENST00000557894.1 ENST00000559284.1 ENST00000560275.1 |
IRF9 |
interferon regulatory factor 9 |
| chr2_-_160473114 | 0.70 |
ENST00000392783.2 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
| chr11_-_89223883 | 0.69 |
ENST00000528341.1 |
NOX4 |
NADPH oxidase 4 |
| chr19_+_10197463 | 0.68 |
ENST00000590378.1 ENST00000397881.3 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr14_-_21270561 | 0.66 |
ENST00000412779.2 |
RNASE1 |
ribonuclease, RNase A family, 1 (pancreatic) |
| chr14_-_100841930 | 0.62 |
ENST00000555031.1 ENST00000553395.1 ENST00000553545.1 ENST00000344102.5 ENST00000556338.1 ENST00000392882.2 ENST00000553934.1 |
WARS |
tryptophanyl-tRNA synthetase |
| chr12_+_58138800 | 0.60 |
ENST00000547992.1 ENST00000552816.1 ENST00000547472.1 |
TSPAN31 |
tetraspanin 31 |
| chr12_+_58138664 | 0.59 |
ENST00000257910.3 |
TSPAN31 |
tetraspanin 31 |
| chr8_-_27469196 | 0.59 |
ENST00000546343.1 ENST00000560566.1 |
CLU |
clusterin |
| chr15_-_60690163 | 0.59 |
ENST00000558998.1 ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2 |
annexin A2 |
| chrX_+_106045891 | 0.58 |
ENST00000357242.5 ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B |
TBC1 domain family, member 8B (with GRAM domain) |
| chr11_+_123986069 | 0.54 |
ENST00000456829.2 ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A |
von Willebrand factor A domain containing 5A |
| chr11_+_33563821 | 0.54 |
ENST00000321505.4 ENST00000265654.5 ENST00000389726.3 |
KIAA1549L |
KIAA1549-like |
| chr14_-_100841670 | 0.51 |
ENST00000557297.1 ENST00000555813.1 ENST00000557135.1 ENST00000556698.1 ENST00000554509.1 ENST00000555410.1 |
WARS |
tryptophanyl-tRNA synthetase |
| chr8_-_95274536 | 0.46 |
ENST00000297596.2 ENST00000396194.2 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
| chr6_+_134210243 | 0.46 |
ENST00000367882.4 |
TCF21 |
transcription factor 21 |
| chr6_+_32821924 | 0.46 |
ENST00000374859.2 ENST00000453265.2 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr11_+_65657875 | 0.45 |
ENST00000312579.2 |
CCDC85B |
coiled-coil domain containing 85B |
| chr8_-_27468842 | 0.42 |
ENST00000523500.1 |
CLU |
clusterin |
| chr15_+_64680003 | 0.42 |
ENST00000261884.3 |
TRIP4 |
thyroid hormone receptor interactor 4 |
| chr2_-_160472952 | 0.42 |
ENST00000541068.2 ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
| chr12_+_56324756 | 0.36 |
ENST00000331886.5 ENST00000555090.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr2_-_175547571 | 0.33 |
ENST00000409415.3 ENST00000359761.3 ENST00000272746.5 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr17_-_77924627 | 0.32 |
ENST00000572862.1 ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16 |
TBC1 domain family, member 16 |
| chr14_+_55034599 | 0.32 |
ENST00000392067.3 ENST00000357634.3 |
SAMD4A |
sterile alpha motif domain containing 4A |
| chr19_-_39881777 | 0.32 |
ENST00000595564.1 ENST00000221265.3 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr12_+_5541267 | 0.31 |
ENST00000423158.3 |
NTF3 |
neurotrophin 3 |
| chr12_+_27396901 | 0.31 |
ENST00000541191.1 ENST00000389032.3 |
STK38L |
serine/threonine kinase 38 like |
| chr10_-_103578182 | 0.31 |
ENST00000439817.1 |
MGEA5 |
meningioma expressed antigen 5 (hyaluronidase) |
| chrX_-_107334790 | 0.30 |
ENST00000217958.3 |
PSMD10 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chrX_-_107334750 | 0.29 |
ENST00000340200.5 ENST00000372296.1 ENST00000372295.1 ENST00000361815.5 |
PSMD10 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr19_-_39881669 | 0.28 |
ENST00000221266.7 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr11_+_6411670 | 0.28 |
ENST00000530395.1 ENST00000527275.1 |
SMPD1 |
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr12_+_60083118 | 0.28 |
ENST00000261187.4 ENST00000543448.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr12_+_26164645 | 0.28 |
ENST00000542004.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr5_+_140227048 | 0.27 |
ENST00000532602.1 |
PCDHA9 |
protocadherin alpha 9 |
| chr11_-_118436707 | 0.26 |
ENST00000264020.2 ENST00000264021.3 |
IFT46 |
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr1_-_9811600 | 0.26 |
ENST00000435891.1 |
CLSTN1 |
calsyntenin 1 |
| chr14_+_96722539 | 0.26 |
ENST00000553356.1 |
BDKRB1 |
bradykinin receptor B1 |
| chr14_+_56127989 | 0.25 |
ENST00000555573.1 |
KTN1 |
kinectin 1 (kinesin receptor) |
| chr17_-_5323480 | 0.25 |
ENST00000573584.1 |
NUP88 |
nucleoporin 88kDa |
| chr13_-_26795840 | 0.24 |
ENST00000381570.3 ENST00000399762.2 ENST00000346166.3 |
RNF6 |
ring finger protein (C3H2C3 type) 6 |
| chr7_-_115670804 | 0.24 |
ENST00000320239.7 |
TFEC |
transcription factor EC |
| chr12_+_53443963 | 0.24 |
ENST00000546602.1 ENST00000552570.1 ENST00000549700.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr13_+_100153665 | 0.23 |
ENST00000376387.4 |
TM9SF2 |
transmembrane 9 superfamily member 2 |
| chr7_-_115670792 | 0.23 |
ENST00000265440.7 ENST00000393485.1 |
TFEC |
transcription factor EC |
| chr5_+_179921344 | 0.22 |
ENST00000261951.4 |
CNOT6 |
CCR4-NOT transcription complex, subunit 6 |
| chr1_+_17906970 | 0.22 |
ENST00000375415.1 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr2_+_210444748 | 0.21 |
ENST00000392194.1 |
MAP2 |
microtubule-associated protein 2 |
| chr12_+_56435637 | 0.20 |
ENST00000356464.5 ENST00000552361.1 |
RPS26 |
ribosomal protein S26 |
| chrX_+_138612889 | 0.19 |
ENST00000218099.2 ENST00000394090.2 |
F9 |
coagulation factor IX |
| chr13_-_41240717 | 0.19 |
ENST00000379561.5 |
FOXO1 |
forkhead box O1 |
| chr3_-_126327398 | 0.18 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr17_-_58603568 | 0.17 |
ENST00000083182.3 |
APPBP2 |
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chrX_-_65253506 | 0.15 |
ENST00000427538.1 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
| chr10_-_116444371 | 0.14 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr6_+_25754927 | 0.14 |
ENST00000377905.4 ENST00000439485.2 |
SLC17A4 |
solute carrier family 17, member 4 |
| chr7_-_82792215 | 0.13 |
ENST00000333891.9 ENST00000423517.2 |
PCLO |
piccolo presynaptic cytomatrix protein |
| chr6_-_49712123 | 0.11 |
ENST00000263045.4 |
CRISP3 |
cysteine-rich secretory protein 3 |
| chr1_-_211752073 | 0.11 |
ENST00000367001.4 |
SLC30A1 |
solute carrier family 30 (zinc transporter), member 1 |
| chr11_-_64490634 | 0.10 |
ENST00000377559.3 ENST00000265459.6 |
NRXN2 |
neurexin 2 |
| chr22_-_50699701 | 0.09 |
ENST00000395780.1 |
MAPK12 |
mitogen-activated protein kinase 12 |
| chr16_+_4674787 | 0.08 |
ENST00000262370.7 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr3_-_57678772 | 0.08 |
ENST00000311128.5 |
DENND6A |
DENN/MADD domain containing 6A |
| chr3_-_194119995 | 0.08 |
ENST00000323007.3 |
GP5 |
glycoprotein V (platelet) |
| chr15_-_75660919 | 0.06 |
ENST00000569482.1 ENST00000565683.1 ENST00000561615.1 ENST00000563622.1 ENST00000568374.1 ENST00000566256.1 ENST00000267978.5 |
MAN2C1 |
mannosidase, alpha, class 2C, member 1 |
| chr22_-_31536480 | 0.06 |
ENST00000215885.3 |
PLA2G3 |
phospholipase A2, group III |
| chr5_-_64777733 | 0.06 |
ENST00000381055.3 |
ADAMTS6 |
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr11_+_128563652 | 0.05 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr1_-_143767881 | 0.05 |
ENST00000419275.1 |
PPIAL4G |
peptidylprolyl isomerase A (cyclophilin A)-like 4G |
| chr7_-_42971759 | 0.05 |
ENST00000538645.1 ENST00000445517.1 ENST00000223321.4 |
PSMA2 |
proteasome (prosome, macropain) subunit, alpha type, 2 |
| chr3_+_122296465 | 0.05 |
ENST00000483793.1 |
PARP15 |
poly (ADP-ribose) polymerase family, member 15 |
| chr12_+_20848282 | 0.04 |
ENST00000545604.1 |
SLCO1C1 |
solute carrier organic anion transporter family, member 1C1 |
| chr11_+_118175132 | 0.04 |
ENST00000361763.4 |
CD3E |
CD3e molecule, epsilon (CD3-TCR complex) |
| chr1_+_9299895 | 0.03 |
ENST00000602477.1 |
H6PD |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr5_-_140013275 | 0.03 |
ENST00000512545.1 ENST00000302014.6 ENST00000401743.2 |
CD14 |
CD14 molecule |
| chr1_-_144364246 | 0.02 |
ENST00000540273.1 |
PPIAL4B |
peptidylprolyl isomerase A (cyclophilin A)-like 4B |
| chr15_+_57998923 | 0.01 |
ENST00000380557.4 |
POLR2M |
polymerase (RNA) II (DNA directed) polypeptide M |
| chr3_-_164875850 | 0.01 |
ENST00000472120.1 |
RP11-747D18.1 |
RP11-747D18.1 |
| chr9_-_116163400 | 0.01 |
ENST00000277315.5 ENST00000448137.1 ENST00000409155.3 |
ALAD |
aminolevulinate dehydratase |
| chr17_+_7358889 | 0.01 |
ENST00000575379.1 |
CHRNB1 |
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr2_+_231921574 | 0.01 |
ENST00000308696.6 ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr9_-_95166884 | 0.01 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr1_-_161519682 | 0.01 |
ENST00000367969.3 ENST00000443193.1 |
FCGR3A |
Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr12_+_56324933 | 0.01 |
ENST00000549629.1 ENST00000555218.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr16_+_58283814 | 0.00 |
ENST00000443128.2 ENST00000219299.4 |
CCDC113 |
coiled-coil domain containing 113 |
| chr6_-_32821599 | 0.00 |
ENST00000354258.4 |
TAP1 |
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.6 | 2.4 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.6 | 1.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 3.5 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.3 | 2.0 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.3 | 1.6 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.3 | 1.0 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 1.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 0.9 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 1.5 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.7 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 1.5 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.3 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 8.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 1.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 3.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 5.9 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.7 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 8.5 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.9 | 3.6 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.8 | 2.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.6 | 1.8 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.5 | 1.6 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.5 | 6.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.4 | 1.9 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.2 | 1.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 2.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 1.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 0.9 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.2 | 1.0 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.2 | 0.5 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.5 | GO:0072275 | branchiomeric skeletal muscle development(GO:0014707) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 1.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.6 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.7 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 1.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 3.5 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 0.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.0 | 1.5 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.3 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.3 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.8 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 1.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 1.4 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 1.8 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 1.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.4 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 8.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.9 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 3.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 2.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.7 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 2.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 4.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 1.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 2.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.6 | 1.9 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.6 | 1.8 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.5 | 8.5 | GO:0032059 | bleb(GO:0032059) |
| 0.3 | 1.6 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 1.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 2.0 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 3.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 1.0 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 2.4 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 5.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 1.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 1.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |