Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
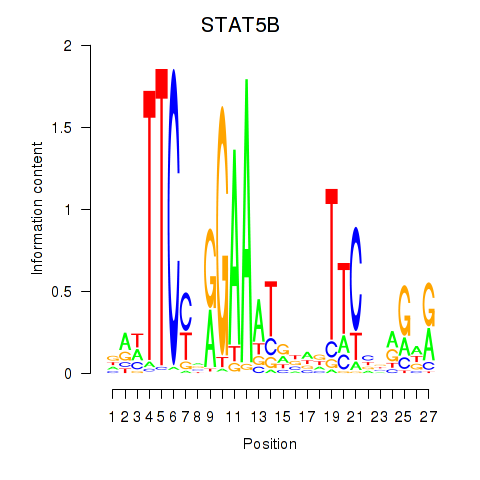
Results for STAT5B
Z-value: 0.88
Transcription factors associated with STAT5B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT5B
|
ENSG00000173757.5 | STAT5B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT5B | hg19_v2_chr17_-_40428359_40428462 | -0.21 | 4.3e-01 | Click! |
Activity profile of STAT5B motif
Sorted Z-values of STAT5B motif
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT5B
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_2709527 | 2.18 |
ENST00000250784.8 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
| chrY_+_2709906 | 2.13 |
ENST00000430575.1 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
| chr5_-_111092930 | 1.88 |
ENST00000257435.7 |
NREP |
neuronal regeneration related protein |
| chr19_+_35168567 | 1.53 |
ENST00000457781.2 ENST00000505163.1 ENST00000505242.1 ENST00000423823.2 ENST00000507959.1 ENST00000446502.2 |
ZNF302 |
zinc finger protein 302 |
| chr5_-_111093167 | 1.50 |
ENST00000446294.2 ENST00000419114.2 |
NREP |
neuronal regeneration related protein |
| chr19_+_35168633 | 1.42 |
ENST00000505365.2 |
ZNF302 |
zinc finger protein 302 |
| chr19_+_54024251 | 1.38 |
ENST00000253144.9 |
ZNF331 |
zinc finger protein 331 |
| chr9_+_112542591 | 1.34 |
ENST00000483909.1 ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2 PALM2-AKAP2 AKAP2 |
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr5_-_111092873 | 1.31 |
ENST00000509025.1 ENST00000515855.1 |
NREP |
neuronal regeneration related protein |
| chr5_-_111093081 | 1.24 |
ENST00000453526.2 ENST00000509427.1 |
NREP |
neuronal regeneration related protein |
| chr5_-_95158644 | 1.17 |
ENST00000237858.6 |
GLRX |
glutaredoxin (thioltransferase) |
| chr5_-_111093340 | 1.06 |
ENST00000508870.1 |
NREP |
neuronal regeneration related protein |
| chr19_+_44669221 | 1.00 |
ENST00000590578.1 ENST00000589160.1 ENST00000337433.5 ENST00000586286.1 ENST00000585560.1 ENST00000586914.1 ENST00000588883.1 ENST00000413984.2 ENST00000588742.1 ENST00000300823.6 ENST00000585678.1 ENST00000586203.1 ENST00000590467.1 ENST00000588795.1 ENST00000588127.1 |
ZNF226 |
zinc finger protein 226 |
| chr12_+_102271129 | 0.94 |
ENST00000258534.8 |
DRAM1 |
DNA-damage regulated autophagy modulator 1 |
| chr5_-_95158375 | 0.84 |
ENST00000512469.2 ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX |
glutaredoxin (thioltransferase) |
| chr19_-_44860820 | 0.83 |
ENST00000354340.4 ENST00000337401.4 ENST00000587909.1 |
ZNF112 |
zinc finger protein 112 |
| chr19_+_44556158 | 0.78 |
ENST00000434772.3 ENST00000585552.1 |
ZNF223 |
zinc finger protein 223 |
| chr18_-_53253323 | 0.76 |
ENST00000540999.1 ENST00000563888.2 |
TCF4 |
transcription factor 4 |
| chr19_+_54041333 | 0.75 |
ENST00000411977.2 ENST00000511154.1 |
ZNF331 |
zinc finger protein 331 |
| chr17_+_32582293 | 0.74 |
ENST00000580907.1 ENST00000225831.4 |
CCL2 |
chemokine (C-C motif) ligand 2 |
| chr12_+_133657461 | 0.72 |
ENST00000412146.2 ENST00000544426.1 ENST00000440984.2 ENST00000319849.3 ENST00000440550.2 |
ZNF140 |
zinc finger protein 140 |
| chr17_+_48624450 | 0.72 |
ENST00000006658.6 ENST00000356488.4 ENST00000393244.3 |
SPATA20 |
spermatogenesis associated 20 |
| chr5_-_42812143 | 0.70 |
ENST00000514985.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr1_+_149871135 | 0.66 |
ENST00000369152.5 |
BOLA1 |
bolA family member 1 |
| chr19_+_56652556 | 0.60 |
ENST00000337080.3 |
ZNF444 |
zinc finger protein 444 |
| chr19_+_56652686 | 0.55 |
ENST00000592949.1 |
ZNF444 |
zinc finger protein 444 |
| chr19_-_46366392 | 0.53 |
ENST00000598059.1 ENST00000594293.1 ENST00000245934.7 |
SYMPK |
symplekin |
| chr4_-_155511887 | 0.52 |
ENST00000302053.3 ENST00000403106.3 |
FGA |
fibrinogen alpha chain |
| chr14_-_70883708 | 0.52 |
ENST00000256366.4 |
SYNJ2BP |
synaptojanin 2 binding protein |
| chrX_-_154250989 | 0.51 |
ENST00000360256.4 |
F8 |
coagulation factor VIII, procoagulant component |
| chr1_+_220921582 | 0.51 |
ENST00000359316.2 |
MARC2 |
mitochondrial amidoxime reducing component 2 |
| chr19_+_44507091 | 0.51 |
ENST00000429154.2 ENST00000585632.1 |
ZNF230 |
zinc finger protein 230 |
| chr11_-_82997013 | 0.46 |
ENST00000529073.1 ENST00000529611.1 |
CCDC90B |
coiled-coil domain containing 90B |
| chrX_+_106045891 | 0.46 |
ENST00000357242.5 ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B |
TBC1 domain family, member 8B (with GRAM domain) |
| chr5_+_178450753 | 0.46 |
ENST00000444149.2 ENST00000519896.1 ENST00000522442.1 |
ZNF879 |
zinc finger protein 879 |
| chr9_+_104161123 | 0.41 |
ENST00000374861.3 ENST00000339664.2 ENST00000259395.4 |
ZNF189 |
zinc finger protein 189 |
| chr19_+_42746927 | 0.40 |
ENST00000378108.1 |
AC006486.1 |
AC006486.1 |
| chr1_+_220921640 | 0.40 |
ENST00000366913.3 |
MARC2 |
mitochondrial amidoxime reducing component 2 |
| chr4_-_89080003 | 0.39 |
ENST00000237612.3 |
ABCG2 |
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr16_-_89785777 | 0.39 |
ENST00000561976.1 |
VPS9D1 |
VPS9 domain containing 1 |
| chr19_+_44598534 | 0.39 |
ENST00000336976.6 |
ZNF224 |
zinc finger protein 224 |
| chr19_+_58038683 | 0.38 |
ENST00000240719.3 ENST00000376233.3 ENST00000594943.1 ENST00000602149.1 |
ZNF549 |
zinc finger protein 549 |
| chr17_-_80291818 | 0.38 |
ENST00000269389.3 ENST00000581691.1 |
SECTM1 |
secreted and transmembrane 1 |
| chr17_-_26697304 | 0.38 |
ENST00000536498.1 |
VTN |
vitronectin |
| chr11_-_82997420 | 0.38 |
ENST00000455220.2 ENST00000529689.1 |
CCDC90B |
coiled-coil domain containing 90B |
| chr14_-_82000140 | 0.37 |
ENST00000555824.1 ENST00000557372.1 ENST00000336735.4 |
SEL1L |
sel-1 suppressor of lin-12-like (C. elegans) |
| chr6_+_139094657 | 0.37 |
ENST00000332797.6 |
CCDC28A |
coiled-coil domain containing 28A |
| chr17_-_34308524 | 0.36 |
ENST00000293275.3 |
CCL16 |
chemokine (C-C motif) ligand 16 |
| chr16_-_18462221 | 0.35 |
ENST00000528301.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr4_+_155484155 | 0.35 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr3_+_14166440 | 0.35 |
ENST00000306077.4 |
TMEM43 |
transmembrane protein 43 |
| chr14_+_23235886 | 0.33 |
ENST00000604262.1 ENST00000431881.2 ENST00000412791.1 ENST00000358043.5 |
OXA1L |
oxidase (cytochrome c) assembly 1-like |
| chr19_+_35225060 | 0.33 |
ENST00000599244.1 ENST00000392232.3 |
ZNF181 |
zinc finger protein 181 |
| chr20_-_43280325 | 0.33 |
ENST00000537820.1 |
ADA |
adenosine deaminase |
| chr19_-_57988871 | 0.32 |
ENST00000596831.1 ENST00000601768.1 ENST00000356584.3 ENST00000600175.1 ENST00000425074.3 ENST00000343280.4 ENST00000427512.2 |
AC004076.9 ZNF772 |
Uncharacterized protein zinc finger protein 772 |
| chr8_+_17780483 | 0.31 |
ENST00000517730.1 ENST00000518537.1 ENST00000523055.1 ENST00000519253.1 |
PCM1 |
pericentriolar material 1 |
| chrX_+_107288239 | 0.31 |
ENST00000217957.5 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr15_+_67420441 | 0.30 |
ENST00000558894.1 |
SMAD3 |
SMAD family member 3 |
| chr7_-_93633684 | 0.30 |
ENST00000222547.3 ENST00000425626.1 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
| chr11_-_82997371 | 0.30 |
ENST00000525503.1 |
CCDC90B |
coiled-coil domain containing 90B |
| chr1_+_54519242 | 0.30 |
ENST00000234827.1 |
TCEANC2 |
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr10_+_93558069 | 0.30 |
ENST00000371627.4 |
TNKS2 |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
| chrX_+_107288197 | 0.30 |
ENST00000415430.3 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr20_+_35973080 | 0.29 |
ENST00000445403.1 |
SRC |
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr2_+_70121075 | 0.29 |
ENST00000409116.1 |
SNRNP27 |
small nuclear ribonucleoprotein 27kDa (U4/U6.U5) |
| chr9_+_130186653 | 0.28 |
ENST00000342483.5 ENST00000543471.1 |
ZNF79 |
zinc finger protein 79 |
| chr7_-_93633658 | 0.28 |
ENST00000433727.1 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
| chr4_+_86396265 | 0.28 |
ENST00000395184.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr2_-_27886676 | 0.27 |
ENST00000337768.5 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr19_+_10197463 | 0.27 |
ENST00000590378.1 ENST00000397881.3 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr19_-_44809121 | 0.26 |
ENST00000591609.1 ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235 |
zinc finger protein 235 |
| chr1_+_149871171 | 0.26 |
ENST00000369150.1 |
BOLA1 |
bolA family member 1 |
| chr5_-_137514288 | 0.25 |
ENST00000454473.1 ENST00000418329.1 ENST00000455658.2 ENST00000230901.5 ENST00000402931.1 |
BRD8 |
bromodomain containing 8 |
| chr4_+_155484103 | 0.25 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr19_+_37341260 | 0.25 |
ENST00000589046.1 |
ZNF345 |
zinc finger protein 345 |
| chr19_+_45445491 | 0.24 |
ENST00000592954.1 ENST00000419266.2 ENST00000589057.1 |
APOC4 APOC4-APOC2 |
apolipoprotein C-IV APOC4-APOC2 readthrough (NMD candidate) |
| chr19_+_44488330 | 0.24 |
ENST00000591532.1 ENST00000407951.2 ENST00000270014.2 ENST00000590615.1 ENST00000586454.1 |
ZNF155 |
zinc finger protein 155 |
| chr7_-_26904317 | 0.24 |
ENST00000345317.2 |
SKAP2 |
src kinase associated phosphoprotein 2 |
| chr5_-_137514333 | 0.23 |
ENST00000411594.2 ENST00000430331.1 |
BRD8 |
bromodomain containing 8 |
| chr17_+_57784826 | 0.23 |
ENST00000262291.4 |
VMP1 |
vacuole membrane protein 1 |
| chr12_-_10324716 | 0.23 |
ENST00000545927.1 ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1 |
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr2_-_63815628 | 0.22 |
ENST00000409562.3 |
WDPCP |
WD repeat containing planar cell polarity effector |
| chr6_+_88182643 | 0.22 |
ENST00000369556.3 ENST00000544441.1 ENST00000369552.4 ENST00000369557.5 |
SLC35A1 |
solute carrier family 35 (CMP-sialic acid transporter), member A1 |
| chr19_+_37341752 | 0.21 |
ENST00000586933.1 ENST00000532141.1 ENST00000420450.1 ENST00000526123.1 |
ZNF345 |
zinc finger protein 345 |
| chr6_+_30130969 | 0.21 |
ENST00000376694.4 |
TRIM15 |
tripartite motif containing 15 |
| chr20_-_25677465 | 0.21 |
ENST00000252979.5 |
ZNF337 |
zinc finger protein 337 |
| chr5_+_178286925 | 0.20 |
ENST00000322434.3 |
ZNF354B |
zinc finger protein 354B |
| chr8_-_146012660 | 0.19 |
ENST00000343459.4 ENST00000429371.2 ENST00000534445.1 |
ZNF34 |
zinc finger protein 34 |
| chr5_-_178157700 | 0.19 |
ENST00000335815.2 |
ZNF354A |
zinc finger protein 354A |
| chr3_-_160823158 | 0.19 |
ENST00000392779.2 ENST00000392780.1 ENST00000494173.1 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr1_-_54519134 | 0.19 |
ENST00000371341.1 |
TMEM59 |
transmembrane protein 59 |
| chr3_-_160823040 | 0.18 |
ENST00000484127.1 ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr2_-_27886460 | 0.18 |
ENST00000404798.2 ENST00000405491.1 ENST00000464789.2 ENST00000406540.1 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr19_-_58220517 | 0.18 |
ENST00000512439.2 ENST00000426889.1 |
ZNF154 |
zinc finger protein 154 |
| chr19_+_39881951 | 0.18 |
ENST00000315588.5 ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29 |
mediator complex subunit 29 |
| chr19_-_36980337 | 0.17 |
ENST00000434377.2 ENST00000424129.2 ENST00000452939.1 ENST00000427002.1 |
ZNF566 |
zinc finger protein 566 |
| chr19_-_50529193 | 0.17 |
ENST00000596445.1 ENST00000599538.1 |
VRK3 |
vaccinia related kinase 3 |
| chr19_-_36980455 | 0.17 |
ENST00000454319.1 ENST00000392170.2 |
ZNF566 |
zinc finger protein 566 |
| chr19_-_56826157 | 0.16 |
ENST00000592509.1 ENST00000592679.1 ENST00000588442.1 ENST00000593106.1 ENST00000587492.1 ENST00000254165.3 |
ZSCAN5A |
zinc finger and SCAN domain containing 5A |
| chr20_-_18774614 | 0.16 |
ENST00000412553.1 |
LINC00652 |
long intergenic non-protein coding RNA 652 |
| chr19_-_58427944 | 0.15 |
ENST00000312026.5 ENST00000536263.1 ENST00000598629.1 ENST00000595559.1 ENST00000597515.1 ENST00000599251.1 ENST00000598526.1 |
ZNF417 |
zinc finger protein 417 |
| chr19_+_44617511 | 0.15 |
ENST00000262894.6 ENST00000588926.1 ENST00000592780.1 |
ZNF225 |
zinc finger protein 225 |
| chr2_+_240684554 | 0.15 |
ENST00000407524.1 |
AC093802.1 |
Uncharacterized protein |
| chr19_+_36208877 | 0.14 |
ENST00000420124.1 ENST00000222270.7 ENST00000341701.1 |
KMT2B |
Histone-lysine N-methyltransferase 2B |
| chr9_+_15422702 | 0.14 |
ENST00000380821.3 ENST00000421710.1 |
SNAPC3 |
small nuclear RNA activating complex, polypeptide 3, 50kDa |
| chr1_-_155112883 | 0.14 |
ENST00000368399.1 ENST00000368400.4 ENST00000341298.3 |
DPM3 |
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr11_+_2920951 | 0.14 |
ENST00000347936.2 |
SLC22A18 |
solute carrier family 22, member 18 |
| chr19_+_57946684 | 0.14 |
ENST00000334181.4 ENST00000415248.1 |
ZNF749 |
zinc finger protein 749 |
| chr3_-_160822858 | 0.14 |
ENST00000488170.1 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr1_+_67673297 | 0.13 |
ENST00000425614.1 ENST00000395227.1 |
IL23R |
interleukin 23 receptor |
| chr1_-_54519067 | 0.13 |
ENST00000452421.1 ENST00000420738.1 ENST00000234831.5 ENST00000440019.1 |
TMEM59 |
transmembrane protein 59 |
| chr1_-_16763911 | 0.13 |
ENST00000375577.1 ENST00000335496.1 |
SPATA21 |
spermatogenesis associated 21 |
| chr1_+_119957554 | 0.13 |
ENST00000543831.1 ENST00000433745.1 ENST00000369416.3 |
HSD3B2 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr19_+_49404041 | 0.13 |
ENST00000263273.5 ENST00000424608.1 |
NUCB1 |
nucleobindin 1 |
| chr20_+_388935 | 0.12 |
ENST00000382181.2 ENST00000400247.3 |
RBCK1 |
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr7_+_47834908 | 0.12 |
ENST00000418326.2 |
C7orf69 |
chromosome 7 open reading frame 69 |
| chr1_-_146054494 | 0.12 |
ENST00000401009.2 |
NBPF11 |
neuroblastoma breakpoint family, member 11 |
| chr1_+_53392901 | 0.11 |
ENST00000371514.3 ENST00000528311.1 ENST00000371509.4 ENST00000407246.2 ENST00000371513.5 |
SCP2 |
sterol carrier protein 2 |
| chr19_-_57183114 | 0.11 |
ENST00000537055.2 ENST00000601659.1 |
ZNF835 |
zinc finger protein 835 |
| chr16_-_53737795 | 0.11 |
ENST00000262135.4 ENST00000564374.1 ENST00000566096.1 |
RPGRIP1L |
RPGRIP1-like |
| chr16_+_16434185 | 0.11 |
ENST00000524823.2 |
AC138969.4 |
Protein PKD1P1 |
| chr6_-_27440837 | 0.11 |
ENST00000211936.6 |
ZNF184 |
zinc finger protein 184 |
| chr8_+_27631903 | 0.10 |
ENST00000305188.8 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr19_+_40502938 | 0.10 |
ENST00000599504.1 ENST00000596894.1 ENST00000601138.1 ENST00000600094.1 ENST00000347077.4 |
ZNF546 |
zinc finger protein 546 |
| chr19_+_58361185 | 0.10 |
ENST00000339656.5 ENST00000423137.1 |
ZNF587 |
zinc finger protein 587 |
| chr5_+_178368186 | 0.10 |
ENST00000320129.3 ENST00000519564.1 |
ZNF454 |
zinc finger protein 454 |
| chr11_-_71639670 | 0.09 |
ENST00000533047.1 ENST00000529844.1 |
RP11-849H4.2 |
Putative short transient receptor potential channel 2-like protein |
| chr11_-_67374177 | 0.09 |
ENST00000333139.3 |
C11orf72 |
chromosome 11 open reading frame 72 |
| chr16_-_53737722 | 0.08 |
ENST00000569716.1 ENST00000562588.1 ENST00000562230.1 ENST00000379925.3 ENST00000563746.1 ENST00000568653.3 |
RPGRIP1L |
RPGRIP1-like |
| chr19_+_44455368 | 0.08 |
ENST00000591168.1 ENST00000587682.1 ENST00000251269.5 |
ZNF221 |
zinc finger protein 221 |
| chr8_-_109260897 | 0.08 |
ENST00000521297.1 ENST00000519030.1 ENST00000521440.1 ENST00000518345.1 ENST00000519627.1 ENST00000220849.5 |
EIF3E |
eukaryotic translation initiation factor 3, subunit E |
| chr19_+_44529479 | 0.08 |
ENST00000587846.1 ENST00000187879.8 ENST00000391960.3 |
ZNF222 |
zinc finger protein 222 |
| chr11_-_71639480 | 0.08 |
ENST00000529513.1 |
RP11-849H4.2 |
Putative short transient receptor potential channel 2-like protein |
| chr2_-_75426826 | 0.08 |
ENST00000305249.5 |
TACR1 |
tachykinin receptor 1 |
| chr19_-_39881777 | 0.07 |
ENST00000595564.1 ENST00000221265.3 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr12_+_120740119 | 0.07 |
ENST00000536460.1 ENST00000202967.4 |
SIRT4 |
sirtuin 4 |
| chr19_+_44716678 | 0.07 |
ENST00000586228.1 ENST00000588219.1 ENST00000313040.7 ENST00000589707.1 ENST00000588394.1 ENST00000589005.1 |
ZNF227 |
zinc finger protein 227 |
| chr19_+_45445524 | 0.06 |
ENST00000591600.1 |
APOC4 |
apolipoprotein C-IV |
| chr19_-_38085633 | 0.06 |
ENST00000593133.1 ENST00000590751.1 ENST00000358744.3 ENST00000328550.2 ENST00000451802.2 |
ZNF571 |
zinc finger protein 571 |
| chr19_-_36980756 | 0.06 |
ENST00000493391.1 |
ZNF566 |
zinc finger protein 566 |
| chr3_+_130745688 | 0.06 |
ENST00000510769.1 ENST00000429253.2 ENST00000356918.4 ENST00000510688.1 ENST00000511262.1 ENST00000383366.4 |
NEK11 |
NIMA-related kinase 11 |
| chr3_-_150920979 | 0.06 |
ENST00000309180.5 ENST00000480322.1 |
GPR171 |
G protein-coupled receptor 171 |
| chr20_+_34824355 | 0.06 |
ENST00000397286.3 ENST00000320849.4 ENST00000373932.3 |
AAR2 |
AAR2 splicing factor homolog (S. cerevisiae) |
| chr11_+_71640112 | 0.05 |
ENST00000530137.1 |
RNF121 |
ring finger protein 121 |
| chr6_-_27440460 | 0.05 |
ENST00000377419.1 |
ZNF184 |
zinc finger protein 184 |
| chr3_+_130745769 | 0.04 |
ENST00000412440.2 |
NEK11 |
NIMA-related kinase 11 |
| chr14_-_20881579 | 0.04 |
ENST00000556935.1 ENST00000262715.5 ENST00000556549.1 |
TEP1 |
telomerase-associated protein 1 |
| chr8_-_56685859 | 0.04 |
ENST00000523423.1 ENST00000523073.1 ENST00000519784.1 ENST00000434581.2 ENST00000519780.1 ENST00000521229.1 ENST00000522576.1 ENST00000523180.1 ENST00000522090.1 |
TMEM68 |
transmembrane protein 68 |
| chr11_+_71640071 | 0.04 |
ENST00000533380.1 ENST00000393713.3 ENST00000545854.1 |
RNF121 |
ring finger protein 121 |
| chr3_+_151531810 | 0.04 |
ENST00000232892.7 |
AADAC |
arylacetamide deacetylase |
| chr6_+_30131318 | 0.03 |
ENST00000376688.1 |
TRIM15 |
tripartite motif containing 15 |
| chr20_+_39657454 | 0.03 |
ENST00000361337.2 |
TOP1 |
topoisomerase (DNA) I |
| chr19_-_44439387 | 0.03 |
ENST00000269973.5 |
ZNF45 |
zinc finger protein 45 |
| chr12_-_110883346 | 0.03 |
ENST00000547365.1 |
ARPC3 |
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr3_-_130745571 | 0.03 |
ENST00000514044.1 ENST00000264992.3 |
ASTE1 |
asteroid homolog 1 (Drosophila) |
| chrX_-_70474910 | 0.02 |
ENST00000373988.1 ENST00000373998.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr4_+_40194609 | 0.02 |
ENST00000508513.1 |
RHOH |
ras homolog family member H |
| chr7_-_108096822 | 0.01 |
ENST00000379028.3 ENST00000413765.2 ENST00000379022.4 |
NRCAM |
neuronal cell adhesion molecule |
| chr7_+_47834880 | 0.01 |
ENST00000258776.4 |
C7orf69 |
chromosome 7 open reading frame 69 |
| chr19_-_40023450 | 0.01 |
ENST00000326282.4 |
EID2B |
EP300 interacting inhibitor of differentiation 2B |
| chr8_-_56685966 | 0.01 |
ENST00000334667.2 |
TMEM68 |
transmembrane protein 68 |
| chr2_+_27886330 | 0.01 |
ENST00000326019.6 |
SLC4A1AP |
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr2_+_17721230 | 0.00 |
ENST00000457525.1 |
VSNL1 |
visinin-like 1 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 0.9 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.2 | 2.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.2 | 0.5 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 4.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0004769 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.4 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.1 | 0.9 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 1.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.3 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.3 | GO:0046102 | hypoxanthine salvage(GO:0043103) inosine metabolic process(GO:0046102) |
| 0.1 | 2.0 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 0.3 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.1 | 0.3 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.2 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.1 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.3 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.2 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.1 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.4 | GO:0015886 | heme transport(GO:0015886) xenobiotic transport(GO:0042908) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.6 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 4.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.5 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) operant conditioning(GO:0035106) |
| 0.0 | 0.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.4 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.7 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.2 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 4.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 6.5 | REACTOME GENERIC TRANSCRIPTION PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 1.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.5 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 0.4 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 4.3 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.6 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 1.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |