Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
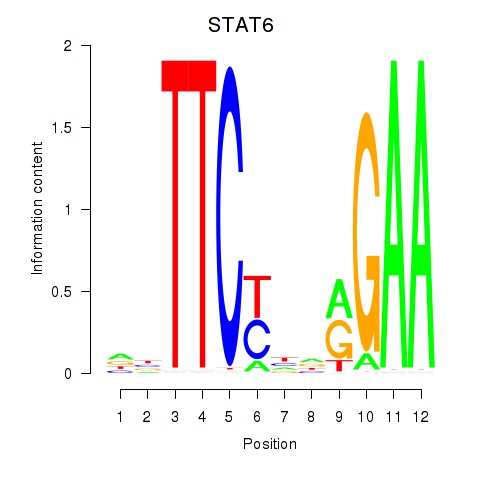
Results for STAT6
Z-value: 1.17
Transcription factors associated with STAT6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT6
|
ENSG00000166888.6 | STAT6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| STAT6 | hg19_v2_chr12_-_57505121_57505216 | 0.65 | 6.2e-03 | Click! |
Activity profile of STAT6 motif
Sorted Z-values of STAT6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of STAT6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_158991025 | 4.05 |
ENST00000337808.6 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr21_+_42539701 | 4.02 |
ENST00000330333.6 ENST00000328735.6 ENST00000347667.5 |
BACE2 |
beta-site APP-cleaving enzyme 2 |
| chr5_+_102201722 | 3.95 |
ENST00000274392.9 ENST00000455264.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr3_+_158787041 | 3.94 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr1_-_68299130 | 3.21 |
ENST00000370982.3 |
GNG12 |
guanine nucleotide binding protein (G protein), gamma 12 |
| chrX_+_135251783 | 2.87 |
ENST00000394153.2 |
FHL1 |
four and a half LIM domains 1 |
| chr2_-_190044480 | 2.85 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr5_+_102201687 | 2.30 |
ENST00000304400.7 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chrX_+_135252050 | 2.06 |
ENST00000449474.1 ENST00000345434.3 |
FHL1 |
four and a half LIM domains 1 |
| chr1_+_150245177 | 1.84 |
ENST00000369098.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr11_-_14379997 | 1.75 |
ENST00000526063.1 ENST00000532814.1 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
| chr13_+_73629107 | 1.67 |
ENST00000539231.1 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chrX_+_135251835 | 1.62 |
ENST00000456445.1 |
FHL1 |
four and a half LIM domains 1 |
| chr17_+_32582293 | 1.56 |
ENST00000580907.1 ENST00000225831.4 |
CCL2 |
chemokine (C-C motif) ligand 2 |
| chr14_-_23285011 | 1.55 |
ENST00000397532.3 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr15_+_25200074 | 1.55 |
ENST00000390687.4 ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr14_-_23285069 | 1.53 |
ENST00000554758.1 ENST00000397528.4 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr1_+_150245099 | 1.51 |
ENST00000369099.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr15_+_25200108 | 1.48 |
ENST00000577949.1 ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF SNRPN |
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr7_-_15726296 | 1.44 |
ENST00000262041.5 |
MEOX2 |
mesenchyme homeobox 2 |
| chr15_-_23932437 | 1.30 |
ENST00000331837.4 |
NDN |
necdin, melanoma antigen (MAGE) family member |
| chr20_-_23066953 | 1.26 |
ENST00000246006.4 |
CD93 |
CD93 molecule |
| chrX_-_106960285 | 1.25 |
ENST00000503515.1 ENST00000372397.2 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr14_-_69446034 | 1.20 |
ENST00000193403.6 |
ACTN1 |
actinin, alpha 1 |
| chr22_+_44464923 | 1.17 |
ENST00000404989.1 |
PARVB |
parvin, beta |
| chr8_+_104311059 | 1.14 |
ENST00000358755.4 ENST00000523739.1 ENST00000540287.1 |
FZD6 |
frizzled family receptor 6 |
| chr3_-_73673991 | 1.12 |
ENST00000308537.4 ENST00000263666.4 |
PDZRN3 |
PDZ domain containing ring finger 3 |
| chr4_-_186877806 | 1.11 |
ENST00000355634.5 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chrX_+_100878079 | 1.11 |
ENST00000471229.2 |
ARMCX3 |
armadillo repeat containing, X-linked 3 |
| chr11_+_77532233 | 1.07 |
ENST00000525409.1 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr5_+_35856951 | 1.06 |
ENST00000303115.3 ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R |
interleukin 7 receptor |
| chr4_+_30721968 | 1.02 |
ENST00000361762.2 |
PCDH7 |
protocadherin 7 |
| chr6_-_11232891 | 0.99 |
ENST00000379433.5 ENST00000379446.5 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
| chr2_-_16804320 | 0.97 |
ENST00000355549.2 |
FAM49A |
family with sequence similarity 49, member A |
| chr2_+_120124497 | 0.97 |
ENST00000355857.3 ENST00000535617.1 ENST00000535757.1 ENST00000409094.1 ENST00000311521.4 |
DBI |
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr2_-_133427767 | 0.96 |
ENST00000397463.2 |
LYPD1 |
LY6/PLAUR domain containing 1 |
| chr8_+_9953061 | 0.90 |
ENST00000522907.1 ENST00000528246.1 |
MSRA |
methionine sulfoxide reductase A |
| chr8_+_9953214 | 0.85 |
ENST00000382490.5 |
MSRA |
methionine sulfoxide reductase A |
| chr10_-_101769617 | 0.84 |
ENST00000324109.4 ENST00000342239.3 |
DNMBP |
dynamin binding protein |
| chr2_+_191792376 | 0.84 |
ENST00000409428.1 ENST00000409215.1 |
GLS |
glutaminase |
| chr19_+_15218180 | 0.79 |
ENST00000342784.2 ENST00000597977.1 ENST00000600440.1 |
SYDE1 |
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr9_-_13175823 | 0.77 |
ENST00000545857.1 |
MPDZ |
multiple PDZ domain protein |
| chr11_+_77532155 | 0.76 |
ENST00000532481.1 ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr11_+_65082289 | 0.75 |
ENST00000279249.2 |
CDC42EP2 |
CDC42 effector protein (Rho GTPase binding) 2 |
| chr12_-_71031185 | 0.73 |
ENST00000548122.1 ENST00000551525.1 ENST00000550358.1 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr14_+_55034599 | 0.69 |
ENST00000392067.3 ENST00000357634.3 |
SAMD4A |
sterile alpha motif domain containing 4A |
| chr2_-_202316260 | 0.68 |
ENST00000332624.3 |
TRAK2 |
trafficking protein, kinesin binding 2 |
| chr3_+_179370517 | 0.62 |
ENST00000263966.3 |
USP13 |
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr8_+_97506033 | 0.61 |
ENST00000518385.1 |
SDC2 |
syndecan 2 |
| chr6_-_24877490 | 0.54 |
ENST00000540914.1 ENST00000378023.4 |
FAM65B |
family with sequence similarity 65, member B |
| chr12_+_12764773 | 0.49 |
ENST00000228865.2 |
CREBL2 |
cAMP responsive element binding protein-like 2 |
| chr17_+_40912764 | 0.49 |
ENST00000589683.1 ENST00000588928.1 |
RAMP2 |
receptor (G protein-coupled) activity modifying protein 2 |
| chr12_-_71031220 | 0.44 |
ENST00000334414.6 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr2_-_230786619 | 0.44 |
ENST00000389045.3 ENST00000409677.1 |
TRIP12 |
thyroid hormone receptor interactor 12 |
| chr1_-_108735440 | 0.42 |
ENST00000370041.4 |
SLC25A24 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr11_+_118230287 | 0.42 |
ENST00000252108.3 ENST00000431736.2 |
UBE4A |
ubiquitination factor E4A |
| chr12_-_111358372 | 0.40 |
ENST00000548438.1 ENST00000228841.8 |
MYL2 |
myosin, light chain 2, regulatory, cardiac, slow |
| chr11_+_112832202 | 0.37 |
ENST00000534015.1 |
NCAM1 |
neural cell adhesion molecule 1 |
| chr12_-_6451235 | 0.36 |
ENST00000440083.2 ENST00000162749.2 |
TNFRSF1A |
tumor necrosis factor receptor superfamily, member 1A |
| chr17_+_68071389 | 0.35 |
ENST00000283936.1 ENST00000392671.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr9_+_90112741 | 0.33 |
ENST00000469640.2 |
DAPK1 |
death-associated protein kinase 1 |
| chr22_-_42342733 | 0.33 |
ENST00000402420.1 |
CENPM |
centromere protein M |
| chr11_+_112832133 | 0.33 |
ENST00000524665.1 |
NCAM1 |
neural cell adhesion molecule 1 |
| chr9_+_90112767 | 0.32 |
ENST00000408954.3 |
DAPK1 |
death-associated protein kinase 1 |
| chr17_+_44588877 | 0.31 |
ENST00000576629.1 |
LRRC37A2 |
leucine rich repeat containing 37, member A2 |
| chr11_-_67980744 | 0.31 |
ENST00000401547.2 ENST00000453170.1 ENST00000304363.4 |
SUV420H1 |
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr14_+_56127989 | 0.27 |
ENST00000555573.1 |
KTN1 |
kinectin 1 (kinesin receptor) |
| chr9_+_90112590 | 0.26 |
ENST00000472284.1 |
DAPK1 |
death-associated protein kinase 1 |
| chr22_+_38201114 | 0.26 |
ENST00000340857.2 |
H1F0 |
H1 histone family, member 0 |
| chr12_+_26164645 | 0.25 |
ENST00000542004.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr10_-_65028817 | 0.24 |
ENST00000542921.1 |
JMJD1C |
jumonji domain containing 1C |
| chr20_-_18477862 | 0.24 |
ENST00000337227.4 |
RBBP9 |
retinoblastoma binding protein 9 |
| chr1_+_89246647 | 0.23 |
ENST00000544045.1 |
PKN2 |
protein kinase N2 |
| chr6_+_26440700 | 0.23 |
ENST00000494393.1 ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
| chr2_-_175462456 | 0.22 |
ENST00000409891.1 ENST00000410117.1 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr11_+_112832090 | 0.22 |
ENST00000533760.1 |
NCAM1 |
neural cell adhesion molecule 1 |
| chr2_+_182756615 | 0.20 |
ENST00000431877.2 ENST00000320370.7 |
SSFA2 |
sperm specific antigen 2 |
| chr1_-_94586651 | 0.19 |
ENST00000535735.1 ENST00000370225.3 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr1_-_241803679 | 0.18 |
ENST00000331838.5 |
OPN3 |
opsin 3 |
| chr9_+_90112117 | 0.17 |
ENST00000358077.5 |
DAPK1 |
death-associated protein kinase 1 |
| chr4_-_10686475 | 0.17 |
ENST00000226951.6 |
CLNK |
cytokine-dependent hematopoietic cell linker |
| chr6_-_32821599 | 0.17 |
ENST00000354258.4 |
TAP1 |
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chrX_-_72434628 | 0.16 |
ENST00000536638.1 ENST00000373517.3 |
NAP1L2 |
nucleosome assembly protein 1-like 2 |
| chr1_-_201915590 | 0.15 |
ENST00000367288.4 |
LMOD1 |
leiomodin 1 (smooth muscle) |
| chr12_+_123949053 | 0.14 |
ENST00000350887.5 |
SNRNP35 |
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr2_-_26700900 | 0.14 |
ENST00000338581.6 ENST00000339598.3 ENST00000402415.3 |
OTOF |
otoferlin |
| chr8_-_82359662 | 0.14 |
ENST00000519260.1 ENST00000256103.2 |
PMP2 |
peripheral myelin protein 2 |
| chr5_+_112227311 | 0.13 |
ENST00000391338.1 |
ZRSR1 |
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 1 |
| chr12_+_6493199 | 0.13 |
ENST00000228918.4 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr22_-_29457832 | 0.13 |
ENST00000216071.4 |
C22orf31 |
chromosome 22 open reading frame 31 |
| chr11_-_10828892 | 0.13 |
ENST00000525681.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr5_+_74011328 | 0.12 |
ENST00000513336.1 |
HEXB |
hexosaminidase B (beta polypeptide) |
| chr7_-_122840015 | 0.12 |
ENST00000194130.2 |
SLC13A1 |
solute carrier family 13 (sodium/sulfate symporter), member 1 |
| chr15_+_96869165 | 0.12 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr12_+_56435637 | 0.12 |
ENST00000356464.5 ENST00000552361.1 |
RPS26 |
ribosomal protein S26 |
| chr15_+_27111510 | 0.11 |
ENST00000335625.5 |
GABRA5 |
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr6_-_27880174 | 0.11 |
ENST00000303324.2 |
OR2B2 |
olfactory receptor, family 2, subfamily B, member 2 |
| chr9_+_36572851 | 0.10 |
ENST00000298048.2 ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK |
maternal embryonic leucine zipper kinase |
| chr7_-_76829125 | 0.09 |
ENST00000248598.5 |
FGL2 |
fibrinogen-like 2 |
| chr8_+_123793633 | 0.09 |
ENST00000314393.4 |
ZHX2 |
zinc fingers and homeoboxes 2 |
| chr20_+_57209828 | 0.08 |
ENST00000598340.1 |
MGC4294 |
HCG1785561; MGC4294 protein; Uncharacterized protein |
| chr7_-_150777920 | 0.08 |
ENST00000353841.2 ENST00000297532.6 |
FASTK |
Fas-activated serine/threonine kinase |
| chr6_+_32821924 | 0.08 |
ENST00000374859.2 ENST00000453265.2 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr14_+_22386325 | 0.07 |
ENST00000390439.2 |
TRAV13-2 |
T cell receptor alpha variable 13-2 |
| chr11_-_60719213 | 0.07 |
ENST00000227880.3 |
SLC15A3 |
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr4_+_95972822 | 0.07 |
ENST00000509540.1 ENST00000440890.2 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr19_+_15052301 | 0.07 |
ENST00000248072.3 |
OR7C2 |
olfactory receptor, family 7, subfamily C, member 2 |
| chr13_-_45915221 | 0.06 |
ENST00000309246.5 ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1 |
tumor protein, translationally-controlled 1 |
| chr19_+_38924316 | 0.06 |
ENST00000355481.4 ENST00000360985.3 ENST00000359596.3 |
RYR1 |
ryanodine receptor 1 (skeletal) |
| chr12_-_10151773 | 0.06 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr22_+_32439019 | 0.06 |
ENST00000266088.4 |
SLC5A1 |
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr7_-_150777949 | 0.05 |
ENST00000482571.1 |
FASTK |
Fas-activated serine/threonine kinase |
| chr11_+_118175132 | 0.05 |
ENST00000361763.4 |
CD3E |
CD3e molecule, epsilon (CD3-TCR complex) |
| chr3_-_49851313 | 0.04 |
ENST00000333486.3 |
UBA7 |
ubiquitin-like modifier activating enzyme 7 |
| chr17_+_7461781 | 0.04 |
ENST00000349228.4 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr11_-_77531858 | 0.03 |
ENST00000360355.2 |
RSF1 |
remodeling and spacing factor 1 |
| chr12_+_9144626 | 0.03 |
ENST00000543895.1 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
| chr16_+_67694849 | 0.03 |
ENST00000602551.1 ENST00000458121.2 ENST00000219255.3 |
PARD6A |
par-6 family cell polarity regulator alpha |
| chr3_+_121311966 | 0.02 |
ENST00000338040.4 |
FBXO40 |
F-box protein 40 |
| chr5_+_140602904 | 0.02 |
ENST00000515856.2 ENST00000239449.4 |
PCDHB14 |
protocadherin beta 14 |
| chr15_+_89402148 | 0.01 |
ENST00000560601.1 |
ACAN |
aggrecan |
| chr14_+_38677123 | 0.01 |
ENST00000267377.2 |
SSTR1 |
somatostatin receptor 1 |
| chr1_-_116383738 | 0.00 |
ENST00000320238.3 |
NHLH2 |
nescient helix loop helix 2 |
| chr1_+_152691998 | 0.00 |
ENST00000368775.2 |
C1orf68 |
chromosome 1 open reading frame 68 |
| chr1_+_174417095 | 0.00 |
ENST00000367685.2 |
GPR52 |
G protein-coupled receptor 52 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 2.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 3.0 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 1.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.5 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 0.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 3.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 7.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.8 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 2.4 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 11.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 1.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 2.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 2.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 3.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 3.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.8 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0001519 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.7 | 2.8 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.5 | 1.6 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.5 | 3.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.5 | 4.0 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.4 | 1.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.3 | 1.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.2 | 0.7 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.2 | 0.8 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 1.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.2 | 1.7 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.2 | 0.8 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.2 | 1.2 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.2 | 1.3 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 0.6 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 | 6.6 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 1.1 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.1 | 0.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 1.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.2 | GO:0002456 | T cell mediated immunity(GO:0002456) |
| 0.1 | 0.4 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 3.2 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.1 | 1.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.1 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.7 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.5 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.4 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.4 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 1.3 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 1.0 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.4 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 1.1 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 2.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 1.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 1.3 | GO:0042116 | macrophage activation(GO:0042116) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.5 | 1.6 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.4 | 1.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.3 | 1.7 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.3 | 3.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.3 | 3.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.2 | 0.6 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 4.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 0.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 1.0 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 1.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 1.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.7 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 1.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.4 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 0.5 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.4 | GO:0015216 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 1.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 1.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 1.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 6.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 2.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 1.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |