Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
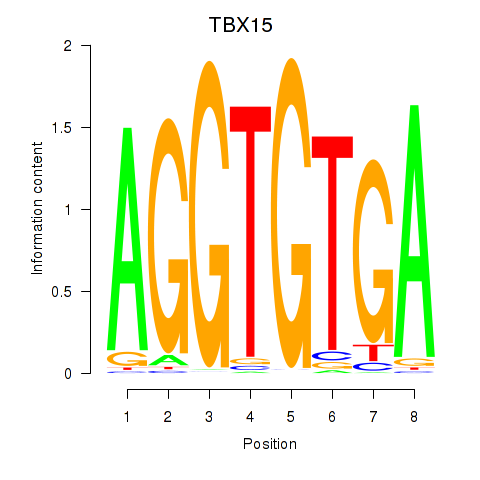
Results for TBX15_MGA
Z-value: 1.27
Transcription factors associated with TBX15_MGA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX15
|
ENSG00000092607.9 | TBX15 |
|
MGA
|
ENSG00000174197.12 | MGA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MGA | hg19_v2_chr15_+_41952591_41952672 | 0.15 | 5.7e-01 | Click! |
Activity profile of TBX15_MGA motif
Sorted Z-values of TBX15_MGA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX15_MGA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_22565101 | 4.79 |
ENST00000419308.2 |
FOXA2 |
forkhead box A2 |
| chr14_+_95078714 | 4.77 |
ENST00000393078.3 ENST00000393080.4 ENST00000467132.1 |
SERPINA3 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr9_-_75567962 | 3.86 |
ENST00000297785.3 ENST00000376939.1 |
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
| chr2_+_228678550 | 3.65 |
ENST00000409189.3 ENST00000358813.4 |
CCL20 |
chemokine (C-C motif) ligand 20 |
| chrX_-_133119476 | 3.31 |
ENST00000543339.1 |
GPC3 |
glypican 3 |
| chr2_-_39664405 | 3.24 |
ENST00000341681.5 ENST00000263881.3 |
MAP4K3 |
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr12_+_56473939 | 2.96 |
ENST00000450146.2 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr6_+_7541808 | 2.81 |
ENST00000379802.3 |
DSP |
desmoplakin |
| chr11_+_73358594 | 2.64 |
ENST00000227214.6 ENST00000398494.4 ENST00000543085.1 |
PLEKHB1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr17_-_1619535 | 2.58 |
ENST00000573075.1 ENST00000574306.1 |
MIR22HG |
MIR22 host gene (non-protein coding) |
| chr2_+_62932779 | 2.48 |
ENST00000427809.1 ENST00000405482.1 ENST00000431489.1 |
EHBP1 |
EH domain binding protein 1 |
| chr11_+_46402297 | 2.43 |
ENST00000405308.2 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr2_+_62933001 | 2.43 |
ENST00000263991.5 ENST00000354487.3 |
EHBP1 |
EH domain binding protein 1 |
| chr6_+_7541845 | 2.31 |
ENST00000418664.2 |
DSP |
desmoplakin |
| chr12_+_69742121 | 2.29 |
ENST00000261267.2 ENST00000549690.1 ENST00000548839.1 |
LYZ |
lysozyme |
| chr17_-_1619491 | 2.27 |
ENST00000570416.1 ENST00000575626.1 ENST00000610106.1 ENST00000608198.1 ENST00000609442.1 ENST00000334146.3 ENST00000576489.1 ENST00000608245.1 ENST00000609398.1 ENST00000608913.1 ENST00000574016.1 ENST00000571091.1 ENST00000573127.1 ENST00000609990.1 ENST00000576749.1 |
MIR22HG |
MIR22 host gene (non-protein coding) |
| chr3_-_120170052 | 2.19 |
ENST00000295633.3 |
FSTL1 |
follistatin-like 1 |
| chrX_-_99891796 | 2.15 |
ENST00000373020.4 |
TSPAN6 |
tetraspanin 6 |
| chr20_+_34680620 | 2.03 |
ENST00000430276.1 ENST00000373950.2 ENST00000452261.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr8_-_27457494 | 2.03 |
ENST00000521770.1 |
CLU |
clusterin |
| chr4_-_186732048 | 1.96 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr11_+_86511569 | 1.89 |
ENST00000441050.1 |
PRSS23 |
protease, serine, 23 |
| chr3_-_49459865 | 1.79 |
ENST00000427987.1 |
AMT |
aminomethyltransferase |
| chr11_-_27494309 | 1.79 |
ENST00000389858.4 |
LGR4 |
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr1_-_161193349 | 1.78 |
ENST00000469730.2 ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2 |
apolipoprotein A-II |
| chr3_+_186330712 | 1.78 |
ENST00000411641.2 ENST00000273784.5 |
AHSG |
alpha-2-HS-glycoprotein |
| chr11_-_27494279 | 1.76 |
ENST00000379214.4 |
LGR4 |
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr10_+_94451574 | 1.74 |
ENST00000492654.2 |
HHEX |
hematopoietically expressed homeobox |
| chr5_-_172756506 | 1.71 |
ENST00000265087.4 |
STC2 |
stanniocalcin 2 |
| chr3_-_99833333 | 1.64 |
ENST00000354552.3 ENST00000331335.5 ENST00000398326.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chr1_-_12677714 | 1.60 |
ENST00000376223.2 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
| chr10_-_15413035 | 1.60 |
ENST00000378116.4 ENST00000455654.1 |
FAM171A1 |
family with sequence similarity 171, member A1 |
| chr3_-_49459878 | 1.60 |
ENST00000546031.1 ENST00000458307.2 ENST00000430521.1 |
AMT |
aminomethyltransferase |
| chr2_-_26101374 | 1.53 |
ENST00000435504.4 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
| chr2_-_175869936 | 1.50 |
ENST00000409900.3 |
CHN1 |
chimerin 1 |
| chr2_-_26101314 | 1.50 |
ENST00000336112.4 ENST00000272341.4 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
| chr2_+_102508955 | 1.49 |
ENST00000414004.2 |
FLJ20373 |
FLJ20373 |
| chrX_+_105937068 | 1.47 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr6_-_31620149 | 1.45 |
ENST00000435080.1 ENST00000375976.4 ENST00000441054.1 |
BAG6 |
BCL2-associated athanogene 6 |
| chr2_-_175870085 | 1.39 |
ENST00000409156.3 |
CHN1 |
chimerin 1 |
| chr6_+_31620191 | 1.36 |
ENST00000375918.2 ENST00000375920.4 |
APOM |
apolipoprotein M |
| chr10_-_92681033 | 1.27 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr2_+_11696464 | 1.26 |
ENST00000234142.5 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chrX_+_70443050 | 1.25 |
ENST00000361726.6 |
GJB1 |
gap junction protein, beta 1, 32kDa |
| chr1_-_226374373 | 1.24 |
ENST00000366812.5 |
ACBD3 |
acyl-CoA binding domain containing 3 |
| chr17_+_57784826 | 1.20 |
ENST00000262291.4 |
VMP1 |
vacuole membrane protein 1 |
| chr12_-_56120838 | 1.17 |
ENST00000548160.1 |
CD63 |
CD63 molecule |
| chr4_-_102268484 | 1.16 |
ENST00000394853.4 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr3_-_66551397 | 1.13 |
ENST00000383703.3 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr11_+_46402482 | 1.13 |
ENST00000441869.1 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr8_+_123793633 | 1.12 |
ENST00000314393.4 |
ZHX2 |
zinc fingers and homeoboxes 2 |
| chr2_+_219283815 | 1.11 |
ENST00000248444.5 ENST00000454069.1 ENST00000392114.2 |
VIL1 |
villin 1 |
| chr1_-_169555779 | 1.08 |
ENST00000367797.3 ENST00000367796.3 |
F5 |
coagulation factor V (proaccelerin, labile factor) |
| chr3_-_66551351 | 1.08 |
ENST00000273261.3 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr1_+_155099927 | 1.05 |
ENST00000368407.3 |
EFNA1 |
ephrin-A1 |
| chr1_+_155100342 | 1.05 |
ENST00000368406.2 |
EFNA1 |
ephrin-A1 |
| chr11_-_6502580 | 1.04 |
ENST00000423813.2 ENST00000396777.3 |
ARFIP2 |
ADP-ribosylation factor interacting protein 2 |
| chr1_+_169077172 | 1.03 |
ENST00000499679.3 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr9_+_75766652 | 1.00 |
ENST00000257497.6 |
ANXA1 |
annexin A1 |
| chr20_-_50385138 | 1.00 |
ENST00000338821.5 |
ATP9A |
ATPase, class II, type 9A |
| chr3_+_142315225 | 0.98 |
ENST00000457734.2 ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1 |
plastin 1 |
| chr4_-_186456766 | 0.98 |
ENST00000284771.6 |
PDLIM3 |
PDZ and LIM domain 3 |
| chr4_-_186456652 | 0.96 |
ENST00000284767.5 ENST00000284770.5 |
PDLIM3 |
PDZ and LIM domain 3 |
| chr11_-_115375107 | 0.95 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chr11_+_61447845 | 0.93 |
ENST00000257215.5 |
DAGLA |
diacylglycerol lipase, alpha |
| chrX_-_48693955 | 0.92 |
ENST00000218230.5 |
PCSK1N |
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr21_+_30502806 | 0.92 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr2_+_128177458 | 0.90 |
ENST00000409048.1 ENST00000422777.3 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr20_+_11898507 | 0.90 |
ENST00000378226.2 |
BTBD3 |
BTB (POZ) domain containing 3 |
| chr3_-_49722523 | 0.88 |
ENST00000448220.1 |
MST1 |
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr17_+_57784997 | 0.87 |
ENST00000537567.1 ENST00000539763.1 ENST00000587945.1 ENST00000536180.1 ENST00000589823.2 ENST00000592106.1 ENST00000591315.1 ENST00000545362.1 |
VMP1 |
vacuole membrane protein 1 |
| chr17_+_4675175 | 0.84 |
ENST00000270560.3 |
TM4SF5 |
transmembrane 4 L six family member 5 |
| chr11_-_6502534 | 0.83 |
ENST00000254584.2 ENST00000525235.1 ENST00000445086.2 |
ARFIP2 |
ADP-ribosylation factor interacting protein 2 |
| chr12_+_104324112 | 0.83 |
ENST00000299767.5 |
HSP90B1 |
heat shock protein 90kDa beta (Grp94), member 1 |
| chr17_-_77924627 | 0.81 |
ENST00000572862.1 ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16 |
TBC1 domain family, member 16 |
| chr16_-_28550320 | 0.81 |
ENST00000395641.2 |
NUPR1 |
nuclear protein, transcriptional regulator, 1 |
| chr2_-_183903133 | 0.80 |
ENST00000361354.4 |
NCKAP1 |
NCK-associated protein 1 |
| chr7_-_76255444 | 0.78 |
ENST00000454397.1 |
POMZP3 |
POM121 and ZP3 fusion |
| chr14_+_55034330 | 0.78 |
ENST00000251091.5 |
SAMD4A |
sterile alpha motif domain containing 4A |
| chr1_+_151584544 | 0.77 |
ENST00000458013.2 ENST00000368843.3 |
SNX27 |
sorting nexin family member 27 |
| chr17_+_35849937 | 0.77 |
ENST00000394389.4 |
DUSP14 |
dual specificity phosphatase 14 |
| chr1_-_19229248 | 0.77 |
ENST00000375341.3 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
| chr4_-_102268628 | 0.75 |
ENST00000323055.6 ENST00000512215.1 ENST00000394854.3 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr6_+_121756809 | 0.74 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chrX_+_135230712 | 0.74 |
ENST00000535737.1 |
FHL1 |
four and a half LIM domains 1 |
| chr14_+_32546274 | 0.74 |
ENST00000396582.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr1_-_19229014 | 0.74 |
ENST00000538839.1 ENST00000290597.5 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
| chr9_+_124048864 | 0.74 |
ENST00000545652.1 |
GSN |
gelsolin |
| chrX_-_131623874 | 0.73 |
ENST00000436215.1 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chr3_+_38017264 | 0.73 |
ENST00000436654.1 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr20_+_4152356 | 0.72 |
ENST00000379460.2 |
SMOX |
spermine oxidase |
| chr16_+_2588012 | 0.71 |
ENST00000354836.5 ENST00000389224.3 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr17_-_57784755 | 0.71 |
ENST00000537860.1 ENST00000393038.2 ENST00000409433.2 |
PTRH2 |
peptidyl-tRNA hydrolase 2 |
| chr12_-_8815299 | 0.69 |
ENST00000535336.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chrX_-_10851762 | 0.68 |
ENST00000380785.1 ENST00000380787.1 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr12_-_8815215 | 0.68 |
ENST00000544889.1 ENST00000543369.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr6_-_160166218 | 0.68 |
ENST00000537657.1 |
SOD2 |
superoxide dismutase 2, mitochondrial |
| chr7_-_27135591 | 0.68 |
ENST00000343060.4 ENST00000355633.5 |
HOXA1 |
homeobox A1 |
| chr11_+_114310102 | 0.67 |
ENST00000265881.5 |
REXO2 |
RNA exonuclease 2 |
| chr1_-_205290865 | 0.67 |
ENST00000367157.3 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
| chrX_-_131623982 | 0.66 |
ENST00000370844.1 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chr18_-_30050395 | 0.66 |
ENST00000269209.6 ENST00000399218.4 |
GAREM |
GRB2 associated, regulator of MAPK1 |
| chr17_-_46682321 | 0.63 |
ENST00000225648.3 ENST00000484302.2 |
HOXB6 |
homeobox B6 |
| chr12_-_56120865 | 0.62 |
ENST00000548898.1 ENST00000552067.1 |
CD63 |
CD63 molecule |
| chr6_-_86099898 | 0.62 |
ENST00000455071.1 |
RP11-30P6.6 |
RP11-30P6.6 |
| chr19_-_48894104 | 0.62 |
ENST00000597017.1 |
KDELR1 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr3_+_52811596 | 0.61 |
ENST00000542827.1 ENST00000273283.2 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr11_+_114310237 | 0.59 |
ENST00000539119.1 |
REXO2 |
RNA exonuclease 2 |
| chr2_-_136288113 | 0.59 |
ENST00000401392.1 |
ZRANB3 |
zinc finger, RAN-binding domain containing 3 |
| chr9_-_73029540 | 0.58 |
ENST00000377126.2 |
KLF9 |
Kruppel-like factor 9 |
| chr19_-_39390350 | 0.58 |
ENST00000447739.1 ENST00000358931.5 ENST00000407552.1 |
SIRT2 |
sirtuin 2 |
| chr17_-_79869004 | 0.58 |
ENST00000573927.1 ENST00000331285.3 ENST00000572157.1 |
PCYT2 |
phosphate cytidylyltransferase 2, ethanolamine |
| chr14_+_55034599 | 0.58 |
ENST00000392067.3 ENST00000357634.3 |
SAMD4A |
sterile alpha motif domain containing 4A |
| chr16_+_69221028 | 0.57 |
ENST00000336278.4 |
SNTB2 |
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2) |
| chr19_+_35773242 | 0.57 |
ENST00000222304.3 |
HAMP |
hepcidin antimicrobial peptide |
| chr9_-_13279563 | 0.56 |
ENST00000541718.1 |
MPDZ |
multiple PDZ domain protein |
| chr17_-_47755338 | 0.56 |
ENST00000508805.1 ENST00000515508.2 ENST00000451526.2 ENST00000507970.1 |
SPOP |
speckle-type POZ protein |
| chr1_-_153066998 | 0.55 |
ENST00000368750.3 |
SPRR2E |
small proline-rich protein 2E |
| chr17_-_79869077 | 0.55 |
ENST00000570391.1 |
PCYT2 |
phosphate cytidylyltransferase 2, ethanolamine |
| chr19_+_39279838 | 0.55 |
ENST00000314980.4 |
LGALS7B |
lectin, galactoside-binding, soluble, 7B |
| chr7_-_6523755 | 0.55 |
ENST00000436575.1 ENST00000258739.4 |
DAGLB KDELR2 |
diacylglycerol lipase, beta KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr14_+_75746781 | 0.55 |
ENST00000555347.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr16_+_85645007 | 0.55 |
ENST00000405402.2 |
GSE1 |
Gse1 coiled-coil protein |
| chr1_-_150780757 | 0.54 |
ENST00000271651.3 |
CTSK |
cathepsin K |
| chr4_-_170924888 | 0.54 |
ENST00000502832.1 ENST00000393704.3 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr14_+_55033815 | 0.53 |
ENST00000554335.1 |
SAMD4A |
sterile alpha motif domain containing 4A |
| chr17_-_10452929 | 0.53 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr19_-_48894762 | 0.53 |
ENST00000600980.1 ENST00000330720.2 |
KDELR1 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr9_-_128003606 | 0.52 |
ENST00000324460.6 |
HSPA5 |
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
| chr17_-_79869243 | 0.52 |
ENST00000538721.2 ENST00000573636.2 ENST00000571105.1 ENST00000576343.1 ENST00000572473.1 |
PCYT2 |
phosphate cytidylyltransferase 2, ethanolamine |
| chrX_+_99899180 | 0.52 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr21_+_35014783 | 0.52 |
ENST00000381291.4 ENST00000381285.4 ENST00000399367.3 ENST00000399352.1 ENST00000399355.2 ENST00000399349.1 |
ITSN1 |
intersectin 1 (SH3 domain protein) |
| chr17_-_79869340 | 0.52 |
ENST00000538936.2 |
PCYT2 |
phosphate cytidylyltransferase 2, ethanolamine |
| chr15_+_90808919 | 0.51 |
ENST00000379095.3 |
NGRN |
neugrin, neurite outgrowth associated |
| chr16_+_28834531 | 0.50 |
ENST00000570200.1 |
ATXN2L |
ataxin 2-like |
| chr14_+_24701819 | 0.50 |
ENST00000560139.1 ENST00000559910.1 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr13_+_115047097 | 0.49 |
ENST00000351487.5 |
UPF3A |
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr1_+_172502336 | 0.49 |
ENST00000263688.3 |
SUCO |
SUN domain containing ossification factor |
| chr4_-_140222358 | 0.48 |
ENST00000505036.1 ENST00000544855.1 ENST00000539002.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr17_-_40333099 | 0.48 |
ENST00000607371.1 |
KCNH4 |
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr19_-_46526304 | 0.47 |
ENST00000008938.4 |
PGLYRP1 |
peptidoglycan recognition protein 1 |
| chr1_+_17906970 | 0.47 |
ENST00000375415.1 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chrX_-_10645773 | 0.46 |
ENST00000453318.2 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr18_+_47088401 | 0.46 |
ENST00000261292.4 ENST00000427224.2 ENST00000580036.1 |
LIPG |
lipase, endothelial |
| chr12_-_8815404 | 0.46 |
ENST00000359478.2 ENST00000396549.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr17_+_39975544 | 0.45 |
ENST00000544340.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr8_-_49834299 | 0.45 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr17_+_39975455 | 0.45 |
ENST00000455106.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr16_+_2587998 | 0.44 |
ENST00000441549.3 ENST00000268673.7 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr19_+_17337473 | 0.44 |
ENST00000598068.1 |
OCEL1 |
occludin/ELL domain containing 1 |
| chr12_-_56121612 | 0.43 |
ENST00000546939.1 |
CD63 |
CD63 molecule |
| chrX_-_153775426 | 0.43 |
ENST00000393562.2 |
G6PD |
glucose-6-phosphate dehydrogenase |
| chr14_+_24701628 | 0.43 |
ENST00000355299.4 ENST00000559836.1 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr2_-_20251744 | 0.42 |
ENST00000175091.4 |
LAPTM4A |
lysosomal protein transmembrane 4 alpha |
| chr14_-_24701539 | 0.41 |
ENST00000534348.1 ENST00000524927.1 ENST00000250495.5 |
NEDD8-MDP1 NEDD8 |
NEDD8-MDP1 readthrough neural precursor cell expressed, developmentally down-regulated 8 |
| chr22_+_35776828 | 0.41 |
ENST00000216117.8 |
HMOX1 |
heme oxygenase (decycling) 1 |
| chr1_-_226129083 | 0.40 |
ENST00000420304.2 |
LEFTY2 |
left-right determination factor 2 |
| chr6_-_30815936 | 0.40 |
ENST00000442852.1 |
XXbac-BPG27H4.8 |
XXbac-BPG27H4.8 |
| chr7_-_71801980 | 0.40 |
ENST00000329008.5 |
CALN1 |
calneuron 1 |
| chr9_-_140082983 | 0.39 |
ENST00000323927.2 |
ANAPC2 |
anaphase promoting complex subunit 2 |
| chr3_+_52821841 | 0.39 |
ENST00000405128.3 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr6_+_43739697 | 0.38 |
ENST00000230480.6 |
VEGFA |
vascular endothelial growth factor A |
| chr3_-_52443799 | 0.38 |
ENST00000470173.1 ENST00000296288.5 |
BAP1 |
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr4_+_90816033 | 0.38 |
ENST00000264790.2 ENST00000394981.1 |
MMRN1 |
multimerin 1 |
| chr12_-_71551868 | 0.38 |
ENST00000247829.3 |
TSPAN8 |
tetraspanin 8 |
| chr12_-_56121580 | 0.38 |
ENST00000550776.1 |
CD63 |
CD63 molecule |
| chr8_-_49833978 | 0.38 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr11_-_66084508 | 0.37 |
ENST00000311330.3 |
CD248 |
CD248 molecule, endosialin |
| chr14_-_24664540 | 0.37 |
ENST00000530563.1 ENST00000528895.1 ENST00000528669.1 ENST00000532632.1 |
TM9SF1 |
transmembrane 9 superfamily member 1 |
| chr1_+_155294342 | 0.37 |
ENST00000292254.4 |
RUSC1 |
RUN and SH3 domain containing 1 |
| chr18_+_60382672 | 0.36 |
ENST00000400316.4 ENST00000262719.5 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
| chr16_+_2587965 | 0.36 |
ENST00000342085.4 ENST00000566659.1 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr19_+_45973120 | 0.36 |
ENST00000592811.1 ENST00000586615.1 |
FOSB |
FBJ murine osteosarcoma viral oncogene homolog B |
| chr14_-_103523745 | 0.35 |
ENST00000361246.2 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
| chrX_-_134049262 | 0.35 |
ENST00000370783.3 |
MOSPD1 |
motile sperm domain containing 1 |
| chr9_+_17134980 | 0.35 |
ENST00000380647.3 |
CNTLN |
centlein, centrosomal protein |
| chr17_-_79869228 | 0.35 |
ENST00000570388.1 |
PCYT2 |
phosphate cytidylyltransferase 2, ethanolamine |
| chr1_+_155294264 | 0.35 |
ENST00000368349.4 |
RUSC1 |
RUN and SH3 domain containing 1 |
| chr1_-_94079648 | 0.35 |
ENST00000370247.3 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr7_-_72972319 | 0.35 |
ENST00000223368.2 |
BCL7B |
B-cell CLL/lymphoma 7B |
| chr17_+_25799008 | 0.34 |
ENST00000583370.1 ENST00000398988.3 ENST00000268763.6 |
KSR1 |
kinase suppressor of ras 1 |
| chrX_+_47077632 | 0.34 |
ENST00000457458.2 |
CDK16 |
cyclin-dependent kinase 16 |
| chr20_+_1115821 | 0.34 |
ENST00000435720.1 |
PSMF1 |
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr20_-_4982132 | 0.33 |
ENST00000338244.1 ENST00000424750.2 |
SLC23A2 |
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr12_-_71551652 | 0.33 |
ENST00000546561.1 |
TSPAN8 |
tetraspanin 8 |
| chr1_+_39456895 | 0.33 |
ENST00000432648.3 ENST00000446189.2 ENST00000372984.4 |
AKIRIN1 |
akirin 1 |
| chr5_+_54320078 | 0.33 |
ENST00000231009.2 |
GZMK |
granzyme K (granzyme 3; tryptase II) |
| chr1_-_52456352 | 0.32 |
ENST00000371655.3 |
RAB3B |
RAB3B, member RAS oncogene family |
| chr6_+_7107999 | 0.32 |
ENST00000491191.1 ENST00000379938.2 ENST00000471433.1 |
RREB1 |
ras responsive element binding protein 1 |
| chr8_-_25281747 | 0.32 |
ENST00000421054.2 |
GNRH1 |
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr6_+_167536230 | 0.32 |
ENST00000341935.5 ENST00000349984.4 |
CCR6 |
chemokine (C-C motif) receptor 6 |
| chr1_-_149889382 | 0.31 |
ENST00000369145.1 ENST00000369146.3 |
SV2A |
synaptic vesicle glycoprotein 2A |
| chr9_+_17135016 | 0.31 |
ENST00000425824.1 ENST00000262360.5 ENST00000380641.4 |
CNTLN |
centlein, centrosomal protein |
| chr9_-_14314566 | 0.31 |
ENST00000397579.2 |
NFIB |
nuclear factor I/B |
| chr1_-_226129189 | 0.31 |
ENST00000366820.5 |
LEFTY2 |
left-right determination factor 2 |
| chr9_-_14314518 | 0.31 |
ENST00000397581.2 |
NFIB |
nuclear factor I/B |
| chr20_+_44509857 | 0.30 |
ENST00000372523.1 ENST00000372520.1 |
ZSWIM1 |
zinc finger, SWIM-type containing 1 |
| chr19_+_54926601 | 0.30 |
ENST00000301194.4 |
TTYH1 |
tweety family member 1 |
| chr17_+_76311791 | 0.30 |
ENST00000586321.1 |
AC061992.2 |
AC061992.2 |
| chr1_+_27114418 | 0.30 |
ENST00000078527.4 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.3 | 4.9 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 4.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 3.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 2.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.8 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.5 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 6.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 2.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 3.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 1.3 | 3.8 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 1.2 | 3.7 | GO:0045360 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.9 | 3.5 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.8 | 3.3 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.7 | 2.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.7 | 5.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.7 | 3.4 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.6 | 1.9 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.6 | 1.8 | GO:0060621 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.6 | 3.6 | GO:0030421 | defecation(GO:0030421) |
| 0.6 | 1.7 | GO:0061010 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.5 | 2.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.5 | 2.3 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.4 | 2.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.4 | 1.5 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.4 | 1.4 | GO:1904327 | maintenance of unfolded protein(GO:0036506) protein localization to cytosolic proteasome complex(GO:1904327) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.3 | 1.4 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.3 | 2.0 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.3 | 1.0 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.3 | 0.9 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.3 | 0.9 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.3 | 3.0 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.3 | 0.8 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.3 | 1.0 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 0.7 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.2 | 2.0 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 1.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 4.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.2 | 0.9 | GO:0099542 | retrograde trans-synaptic signaling(GO:0098917) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.2 | 0.5 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.2 | 1.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.2 | 0.6 | GO:2000777 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.2 | 2.3 | GO:0035437 | maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.2 | 0.6 | GO:1904479 | cellular response to bile acid(GO:1903413) negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 0.2 | 2.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 1.5 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.2 | 0.3 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.2 | 0.5 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.2 | 1.5 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.7 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 1.9 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 0.4 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 3.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) |
| 0.1 | 0.7 | GO:0003070 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.4 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.1 | 0.8 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 0.4 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.4 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 2.7 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 0.7 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.1 | 0.6 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.5 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 2.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 1.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.8 | GO:0001767 | establishment of lymphocyte polarity(GO:0001767) |
| 0.1 | 0.3 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.1 | 0.5 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 0.3 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.1 | 0.1 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 0.2 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.1 | 0.6 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.2 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 | 1.1 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 0.3 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 1.8 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 1.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.8 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.6 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.2 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.3 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.1 | 1.3 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.1 | 2.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.8 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 3.7 | GO:0021545 | cranial nerve development(GO:0021545) |
| 0.1 | 0.8 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.3 | GO:0051582 | peptidyl-cysteine methylation(GO:0018125) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.1 | 0.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.2 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.1 | 2.0 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.9 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 1.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.3 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 0.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0090035 | regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 | 0.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.7 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.2 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.1 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.4 | GO:0002911 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.3 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 1.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 1.0 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 2.1 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 2.6 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.5 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 1.3 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 1.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.9 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.6 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0060969 | negative regulation of gene silencing(GO:0060969) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0060579 | ventral spinal cord interneuron specification(GO:0021521) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) cell fate specification involved in pattern specification(GO:0060573) ventral spinal cord interneuron fate commitment(GO:0060579) cell fate commitment involved in pattern specification(GO:0060581) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.4 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0039003 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) thyroid-stimulating hormone secretion(GO:0070460) kidney rudiment formation(GO:0072003) kidney field specification(GO:0072004) metanephric nephron tubule formation(GO:0072289) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.8 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.4 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.5 | GO:0032967 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.0 | 0.3 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 1.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.4 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.0 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.0 | 0.1 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.1 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0043383 | negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.0 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.1 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.8 | 3.4 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.8 | 3.3 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.8 | 3.9 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.6 | 5.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.5 | 1.5 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.5 | 2.5 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.4 | 1.8 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.4 | 1.8 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.4 | 1.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.4 | 2.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.3 | 0.6 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.2 | 0.7 | GO:0052895 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.2 | 0.7 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 3.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 3.0 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 4.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.2 | 3.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 1.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 0.5 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 0.9 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.7 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 1.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 0.3 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 5.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.9 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 0.8 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.4 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.7 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 0.3 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 1.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 1.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 1.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 1.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 5.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.8 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 1.0 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 1.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 1.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 4.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.6 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 2.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 1.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.4 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.1 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 1.6 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.0 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.0 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.4 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 2.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 2.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 5.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 4.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 1.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 3.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 2.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 2.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 2.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 2.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 4.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.1 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 4.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.4 | 5.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.4 | 1.4 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.3 | 3.8 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 2.6 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.2 | 0.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 1.0 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 5.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 1.0 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 0.9 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.9 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.8 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 1.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.0 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.1 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.1 | 1.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.5 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.1 | 1.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 2.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.5 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 7.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.3 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.5 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.1 | 0.6 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 1.0 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.4 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.3 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.2 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.2 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.7 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 7.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 2.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 1.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 3.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 2.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |