Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for TBX20
Z-value: 0.75
Transcription factors associated with TBX20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX20
|
ENSG00000164532.10 | TBX20 |
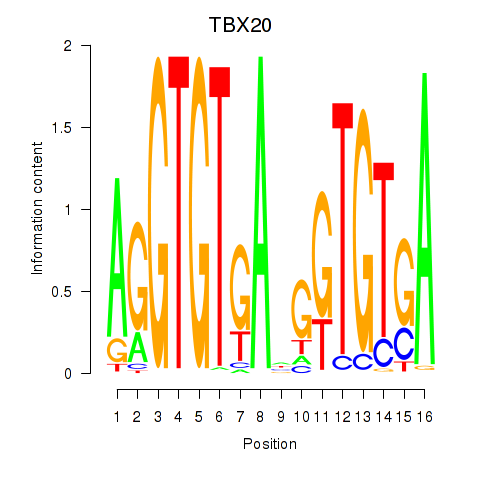
Activity profile of TBX20 motif
Sorted Z-values of TBX20 motif
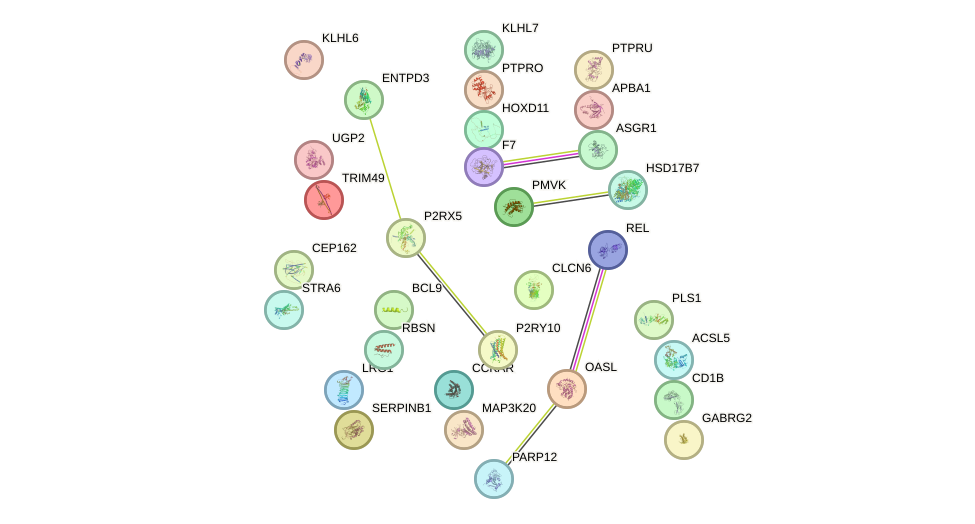
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX20
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_57662419 | 2.91 |
ENST00000388812.4 ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56 |
G protein-coupled receptor 56 |
| chr17_-_7080227 | 0.99 |
ENST00000574330.1 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr10_+_114135004 | 0.89 |
ENST00000393081.1 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
| chr10_+_114135952 | 0.77 |
ENST00000356116.1 ENST00000433418.1 ENST00000354273.4 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
| chr3_+_142315225 | 0.74 |
ENST00000457734.2 ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1 |
plastin 1 |
| chrX_+_78200913 | 0.74 |
ENST00000171757.2 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
| chrX_+_78200829 | 0.73 |
ENST00000544091.1 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
| chr2_+_61108650 | 0.69 |
ENST00000295025.8 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr19_+_56915668 | 0.53 |
ENST00000333201.9 ENST00000391778.3 |
ZNF583 |
zinc finger protein 583 |
| chr19_-_4540486 | 0.51 |
ENST00000306390.6 |
LRG1 |
leucine-rich alpha-2-glycoprotein 1 |
| chr2_+_61108771 | 0.49 |
ENST00000394479.3 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr1_+_162760513 | 0.46 |
ENST00000367915.1 ENST00000367917.3 ENST00000254521.3 ENST00000367913.1 |
HSD17B7 |
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr17_-_3599327 | 0.46 |
ENST00000551178.1 ENST00000552276.1 ENST00000547178.1 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr3_+_40428647 | 0.40 |
ENST00000301825.3 ENST00000439533.1 ENST00000456402.1 |
ENTPD3 |
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr17_-_3599696 | 0.38 |
ENST00000225328.5 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr17_-_3599492 | 0.37 |
ENST00000435558.1 ENST00000345901.3 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr15_-_22448819 | 0.37 |
ENST00000604066.1 |
IGHV1OR15-1 |
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr8_+_27183033 | 0.36 |
ENST00000420218.2 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr8_+_27182862 | 0.35 |
ENST00000521164.1 ENST00000346049.5 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr13_+_113760098 | 0.35 |
ENST00000346342.3 ENST00000541084.1 ENST00000375581.3 |
F7 |
coagulation factor VII (serum prothrombin conversion accelerator) |
| chr1_+_43824669 | 0.34 |
ENST00000372462.1 |
CDC20 |
cell division cycle 20 |
| chr6_-_136788001 | 0.32 |
ENST00000544465.1 |
MAP7 |
microtubule-associated protein 7 |
| chr5_+_133861339 | 0.29 |
ENST00000282605.4 ENST00000361895.2 ENST00000402835.1 |
PHF15 |
jade family PHD finger 2 |
| chr2_+_173940442 | 0.26 |
ENST00000409176.2 ENST00000338983.3 ENST00000431503.2 |
MLTK |
Mitogen-activated protein kinase kinase kinase MLT |
| chr6_-_84937314 | 0.26 |
ENST00000257766.4 ENST00000403245.3 |
KIAA1009 |
KIAA1009 |
| chr12_+_15475331 | 0.25 |
ENST00000281171.4 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr2_+_207630081 | 0.25 |
ENST00000236980.6 ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2 |
FAST kinase domains 2 |
| chr12_+_15475462 | 0.25 |
ENST00000543886.1 ENST00000348962.2 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr9_-_72287191 | 0.21 |
ENST00000265381.4 |
APBA1 |
amyloid beta (A4) precursor protein-binding, family A, member 1 |
| chr15_-_74504560 | 0.20 |
ENST00000449139.2 |
STRA6 |
stimulated by retinoic acid 6 |
| chr15_-_74504597 | 0.20 |
ENST00000416286.3 |
STRA6 |
stimulated by retinoic acid 6 |
| chr2_+_64068074 | 0.19 |
ENST00000394417.2 ENST00000484142.1 ENST00000482668.1 ENST00000467648.2 |
UGP2 |
UDP-glucose pyrophosphorylase 2 |
| chr5_+_161494521 | 0.18 |
ENST00000356592.3 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr22_-_46283597 | 0.17 |
ENST00000451118.1 |
WI2-85898F10.1 |
WI2-85898F10.1 |
| chr14_-_106453155 | 0.16 |
ENST00000390594.2 |
IGHV1-2 |
immunoglobulin heavy variable 1-2 |
| chr2_-_219134822 | 0.15 |
ENST00000444053.1 ENST00000248450.4 |
AAMP |
angio-associated, migratory cell protein |
| chr20_-_48782639 | 0.15 |
ENST00000435301.2 |
RP11-112L6.3 |
RP11-112L6.3 |
| chr20_+_44509857 | 0.14 |
ENST00000372523.1 ENST00000372520.1 |
ZSWIM1 |
zinc finger, SWIM-type containing 1 |
| chr15_-_38519066 | 0.13 |
ENST00000561320.1 ENST00000561161.1 |
RP11-346D14.1 |
RP11-346D14.1 |
| chr6_-_2842219 | 0.12 |
ENST00000380739.5 |
SERPINB1 |
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr2_-_219134343 | 0.12 |
ENST00000447885.1 ENST00000420660.1 |
AAMP |
angio-associated, migratory cell protein |
| chr4_-_26492076 | 0.10 |
ENST00000295589.3 |
CCKAR |
cholecystokinin A receptor |
| chr3_-_15140629 | 0.10 |
ENST00000507357.1 ENST00000449050.1 ENST00000253699.3 ENST00000435849.3 ENST00000476527.2 |
ZFYVE20 |
zinc finger, FYVE domain containing 20 |
| chr1_+_147013182 | 0.10 |
ENST00000234739.3 |
BCL9 |
B-cell CLL/lymphoma 9 |
| chr6_-_2842087 | 0.10 |
ENST00000537185.1 |
SERPINB1 |
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr12_-_121477039 | 0.08 |
ENST00000257570.5 |
OASL |
2'-5'-oligoadenylate synthetase-like |
| chr11_+_6281036 | 0.08 |
ENST00000532715.1 ENST00000525014.1 ENST00000531712.1 ENST00000525462.1 |
CCKBR |
cholecystokinin B receptor |
| chr1_-_11865982 | 0.07 |
ENST00000418034.1 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr2_+_176972000 | 0.07 |
ENST00000249504.5 |
HOXD11 |
homeobox D11 |
| chr7_-_139763521 | 0.06 |
ENST00000263549.3 |
PARP12 |
poly (ADP-ribose) polymerase family, member 12 |
| chr3_-_183273477 | 0.04 |
ENST00000341319.3 |
KLHL6 |
kelch-like family member 6 |
| chr1_-_154909329 | 0.03 |
ENST00000368467.3 |
PMVK |
phosphomevalonate kinase |
| chr1_-_158300747 | 0.03 |
ENST00000451207.1 |
CD1B |
CD1b molecule |
| chr7_-_142207004 | 0.03 |
ENST00000426318.2 |
TRBV10-2 |
T cell receptor beta variable 10-2 |
| chr11_-_89540388 | 0.02 |
ENST00000532501.2 |
TRIM49 |
tripartite motif containing 49 |
| chr1_+_11866207 | 0.02 |
ENST00000312413.6 ENST00000346436.6 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.2 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.2 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:2000538 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.2 | 0.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.3 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 2.7 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.1 | 0.4 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 1.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 1.7 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.2 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.1 | 0.4 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 1.0 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.5 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 0.5 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 1.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 1.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 1.7 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.2 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.2 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 0.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 1.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.5 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.4 | GO:0051183 | vitamin transporter activity(GO:0051183) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |