Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
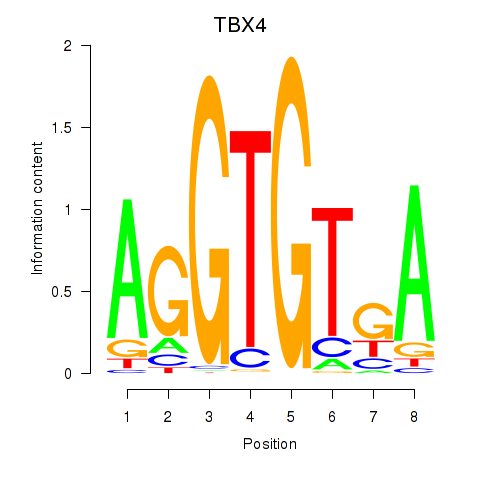
Results for TBX4
Z-value: 0.69
Transcription factors associated with TBX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX4
|
ENSG00000121075.5 | TBX4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX4 | hg19_v2_chr17_+_59529743_59529798 | -0.18 | 4.9e-01 | Click! |
Activity profile of TBX4 motif
Sorted Z-values of TBX4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_75567962 | 2.58 |
ENST00000297785.3 ENST00000376939.1 |
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
| chr2_-_175870085 | 2.34 |
ENST00000409156.3 |
CHN1 |
chimerin 1 |
| chr2_-_175869936 | 2.21 |
ENST00000409900.3 |
CHN1 |
chimerin 1 |
| chr4_-_186456652 | 1.66 |
ENST00000284767.5 ENST00000284770.5 |
PDLIM3 |
PDZ and LIM domain 3 |
| chr4_-_186456766 | 1.65 |
ENST00000284771.6 |
PDLIM3 |
PDZ and LIM domain 3 |
| chr10_+_31608054 | 1.46 |
ENST00000320985.10 ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr4_-_186732048 | 1.24 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr12_-_90024360 | 1.16 |
ENST00000393164.2 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr21_+_30502806 | 1.11 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr5_-_172756506 | 1.05 |
ENST00000265087.4 |
STC2 |
stanniocalcin 2 |
| chr5_+_82767284 | 0.92 |
ENST00000265077.3 |
VCAN |
versican |
| chr5_+_82767487 | 0.85 |
ENST00000343200.5 ENST00000342785.4 |
VCAN |
versican |
| chr6_+_39760129 | 0.73 |
ENST00000274867.4 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr13_-_36429763 | 0.71 |
ENST00000379893.1 |
DCLK1 |
doublecortin-like kinase 1 |
| chr16_+_85645007 | 0.64 |
ENST00000405402.2 |
GSE1 |
Gse1 coiled-coil protein |
| chr11_-_85779971 | 0.60 |
ENST00000393346.3 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
| chr1_-_230561475 | 0.59 |
ENST00000391860.1 |
PGBD5 |
piggyBac transposable element derived 5 |
| chrX_-_11445856 | 0.46 |
ENST00000380736.1 |
ARHGAP6 |
Rho GTPase activating protein 6 |
| chr9_+_34990219 | 0.39 |
ENST00000541010.1 ENST00000454002.2 ENST00000545841.1 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr18_-_53068911 | 0.38 |
ENST00000537856.3 |
TCF4 |
transcription factor 4 |
| chr10_-_75226166 | 0.34 |
ENST00000544628.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr11_+_1940786 | 0.32 |
ENST00000278317.6 ENST00000381561.4 ENST00000381548.3 ENST00000360603.3 ENST00000381549.3 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
| chr1_+_114471972 | 0.29 |
ENST00000369559.4 ENST00000369554.2 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr15_+_89402148 | 0.28 |
ENST00000560601.1 |
ACAN |
aggrecan |
| chr11_+_61447845 | 0.28 |
ENST00000257215.5 |
DAGLA |
diacylglycerol lipase, alpha |
| chr1_+_114472222 | 0.28 |
ENST00000369558.1 ENST00000369561.4 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr15_+_92937058 | 0.22 |
ENST00000268164.3 |
ST8SIA2 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr6_-_31080336 | 0.19 |
ENST00000259870.3 |
C6orf15 |
chromosome 6 open reading frame 15 |
| chr2_-_157189180 | 0.19 |
ENST00000539077.1 ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
| chr15_+_92937144 | 0.18 |
ENST00000539113.1 ENST00000555434.1 |
ST8SIA2 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr1_+_114472481 | 0.17 |
ENST00000369555.2 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr5_-_114515734 | 0.16 |
ENST00000514154.1 ENST00000282369.3 |
TRIM36 |
tripartite motif containing 36 |
| chr15_-_58357932 | 0.16 |
ENST00000347587.3 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr11_+_4116054 | 0.16 |
ENST00000423050.2 |
RRM1 |
ribonucleotide reductase M1 |
| chr10_+_94833642 | 0.15 |
ENST00000224356.4 ENST00000394139.1 |
CYP26A1 |
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr7_-_150675372 | 0.15 |
ENST00000262186.5 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr17_-_8066843 | 0.15 |
ENST00000404970.3 |
VAMP2 |
vesicle-associated membrane protein 2 (synaptobrevin 2) |
| chr9_-_74979420 | 0.15 |
ENST00000343431.2 ENST00000376956.3 |
ZFAND5 |
zinc finger, AN1-type domain 5 |
| chr5_-_60240858 | 0.15 |
ENST00000426742.2 ENST00000265038.5 ENST00000543101.1 ENST00000439176.1 |
ERCC8 |
excision repair cross-complementing rodent repair deficiency, complementation group 8 |
| chr1_+_114471809 | 0.15 |
ENST00000426820.2 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr8_+_123793633 | 0.14 |
ENST00000314393.4 |
ZHX2 |
zinc fingers and homeoboxes 2 |
| chr18_+_32073253 | 0.14 |
ENST00000283365.9 ENST00000596745.1 ENST00000315456.6 |
DTNA |
dystrobrevin, alpha |
| chr5_-_54281407 | 0.14 |
ENST00000381403.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr11_+_73358594 | 0.13 |
ENST00000227214.6 ENST00000398494.4 ENST00000543085.1 |
PLEKHB1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr17_-_61988556 | 0.13 |
ENST00000309894.5 ENST00000438387.2 ENST00000346606.6 ENST00000561003.1 ENST00000450719.3 ENST00000259003.10 |
CSHL1 |
chorionic somatomammotropin hormone-like 1 |
| chr8_-_95907423 | 0.13 |
ENST00000396133.3 ENST00000308108.4 |
CCNE2 |
cyclin E2 |
| chr17_-_26220366 | 0.13 |
ENST00000460380.2 ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9 RP1-66C13.4 |
LYR motif containing 9 Uncharacterized protein |
| chr1_-_1009683 | 0.12 |
ENST00000453464.2 |
RNF223 |
ring finger protein 223 |
| chr2_-_85895295 | 0.12 |
ENST00000428225.1 ENST00000519937.2 |
SFTPB |
surfactant protein B |
| chr16_+_2588012 | 0.12 |
ENST00000354836.5 ENST00000389224.3 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr11_-_5248294 | 0.09 |
ENST00000335295.4 |
HBB |
hemoglobin, beta |
| chr7_+_143013198 | 0.09 |
ENST00000343257.2 |
CLCN1 |
chloride channel, voltage-sensitive 1 |
| chr14_+_78870030 | 0.09 |
ENST00000553631.1 ENST00000554719.1 |
NRXN3 |
neurexin 3 |
| chr20_-_22565101 | 0.09 |
ENST00000419308.2 |
FOXA2 |
forkhead box A2 |
| chr1_+_145507587 | 0.09 |
ENST00000330165.8 ENST00000369307.3 |
RBM8A |
RNA binding motif protein 8A |
| chr11_-_6502580 | 0.08 |
ENST00000423813.2 ENST00000396777.3 |
ARFIP2 |
ADP-ribosylation factor interacting protein 2 |
| chr11_-_6502534 | 0.08 |
ENST00000254584.2 ENST00000525235.1 ENST00000445086.2 |
ARFIP2 |
ADP-ribosylation factor interacting protein 2 |
| chr9_+_71986182 | 0.08 |
ENST00000303068.7 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr5_-_78809950 | 0.08 |
ENST00000334082.6 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr14_-_45603657 | 0.07 |
ENST00000396062.3 |
FKBP3 |
FK506 binding protein 3, 25kDa |
| chr11_-_128737259 | 0.07 |
ENST00000440599.2 ENST00000392666.1 ENST00000324036.3 |
KCNJ1 |
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr16_+_2587965 | 0.07 |
ENST00000342085.4 ENST00000566659.1 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr1_+_95582881 | 0.07 |
ENST00000370203.4 ENST00000456991.1 |
TMEM56 |
transmembrane protein 56 |
| chr3_-_49459865 | 0.07 |
ENST00000427987.1 |
AMT |
aminomethyltransferase |
| chr2_-_111291587 | 0.07 |
ENST00000437167.1 |
RGPD6 |
RANBP2-like and GRIP domain containing 6 |
| chr12_+_50451331 | 0.07 |
ENST00000228468.4 |
ASIC1 |
acid-sensing (proton-gated) ion channel 1 |
| chr11_+_114271251 | 0.06 |
ENST00000375490.5 |
RBM7 |
RNA binding motif protein 7 |
| chr11_+_74660278 | 0.06 |
ENST00000263672.6 ENST00000530257.1 ENST00000526361.1 ENST00000532972.1 |
SPCS2 |
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr5_+_68788594 | 0.06 |
ENST00000396442.2 ENST00000380766.2 |
OCLN |
occludin |
| chr17_-_39222131 | 0.06 |
ENST00000394015.2 |
KRTAP2-4 |
keratin associated protein 2-4 |
| chr11_+_114270752 | 0.06 |
ENST00000540163.1 |
RBM7 |
RNA binding motif protein 7 |
| chr12_+_57853918 | 0.06 |
ENST00000532291.1 ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1 |
GLI family zinc finger 1 |
| chr11_-_128737163 | 0.06 |
ENST00000324003.3 ENST00000392665.2 |
KCNJ1 |
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr11_-_33913708 | 0.05 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr13_+_115047097 | 0.05 |
ENST00000351487.5 |
UPF3A |
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr1_-_111148241 | 0.05 |
ENST00000440270.1 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr17_-_61996160 | 0.05 |
ENST00000458650.2 ENST00000351388.4 ENST00000323322.5 |
GH1 |
growth hormone 1 |
| chr2_+_37458776 | 0.05 |
ENST00000002125.4 ENST00000336237.6 ENST00000431821.1 |
NDUFAF7 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr5_+_113697983 | 0.05 |
ENST00000264773.3 |
KCNN2 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr17_+_30771279 | 0.05 |
ENST00000261712.3 ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr17_-_61996136 | 0.04 |
ENST00000342364.4 |
GH1 |
growth hormone 1 |
| chr3_-_49459878 | 0.04 |
ENST00000546031.1 ENST00000458307.2 ENST00000430521.1 |
AMT |
aminomethyltransferase |
| chrX_+_46771711 | 0.03 |
ENST00000424392.1 ENST00000397189.1 |
PHF16 |
jade family PHD finger 3 |
| chr7_+_97361218 | 0.03 |
ENST00000319273.5 |
TAC1 |
tachykinin, precursor 1 |
| chr5_-_74807418 | 0.03 |
ENST00000405807.4 ENST00000261415.7 |
COL4A3BP |
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chr2_+_11696464 | 0.03 |
ENST00000234142.5 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr11_+_111749942 | 0.03 |
ENST00000260276.3 |
C11orf1 |
chromosome 11 open reading frame 1 |
| chr17_-_61996192 | 0.03 |
ENST00000392824.4 |
CSHL1 |
chorionic somatomammotropin hormone-like 1 |
| chr7_+_97361388 | 0.02 |
ENST00000350485.4 ENST00000346867.4 |
TAC1 |
tachykinin, precursor 1 |
| chr9_-_74980113 | 0.02 |
ENST00000376962.5 ENST00000376960.4 ENST00000237937.3 |
ZFAND5 |
zinc finger, AN1-type domain 5 |
| chr16_+_2587998 | 0.02 |
ENST00000441549.3 ENST00000268673.7 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr5_-_54281491 | 0.02 |
ENST00000381405.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr17_+_57970469 | 0.02 |
ENST00000443572.2 ENST00000406116.3 ENST00000225577.4 ENST00000393021.3 |
RPS6KB1 |
ribosomal protein S6 kinase, 70kDa, polypeptide 1 |
| chr17_-_61973929 | 0.02 |
ENST00000329882.8 ENST00000453363.3 ENST00000316193.8 |
CSH1 |
chorionic somatomammotropin hormone 1 (placental lactogen) |
| chrX_+_46771848 | 0.02 |
ENST00000218343.4 |
PHF16 |
jade family PHD finger 3 |
| chr10_-_99531709 | 0.01 |
ENST00000266066.3 |
SFRP5 |
secreted frizzled-related protein 5 |
| chr3_+_52811596 | 0.01 |
ENST00000542827.1 ENST00000273283.2 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr19_+_10131437 | 0.01 |
ENST00000587782.1 |
RDH8 |
retinol dehydrogenase 8 (all-trans) |
| chr3_+_120626919 | 0.00 |
ENST00000273666.6 ENST00000471454.1 ENST00000472879.1 ENST00000497029.1 ENST00000492541.1 |
STXBP5L |
syntaxin binding protein 5-like |
| chr5_-_34043310 | 0.00 |
ENST00000231338.7 |
C1QTNF3 |
C1q and tumor necrosis factor related protein 3 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 5.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 1.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.2 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 3.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 2.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 1.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.2 | 1.5 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 1.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 4.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.9 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.6 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 1.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.3 | GO:0099542 | retrograde trans-synaptic signaling(GO:0098917) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.1 | 0.2 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.2 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.2 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.1 | GO:2000543 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) positive regulation of gastrulation(GO:2000543) |
| 0.0 | 1.0 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.0 | 0.2 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.0 | 0.5 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.0 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 1.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0007418 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.0 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.0 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.0 | 0.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 4.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 2.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.2 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.2 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 1.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 1.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 3.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.0 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |