Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
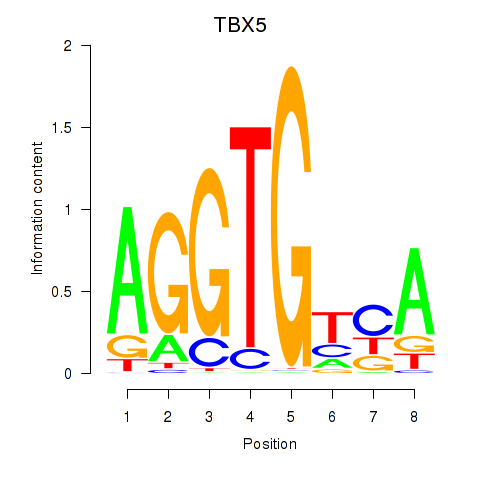
Results for TBX5
Z-value: 0.88
Transcription factors associated with TBX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX5
|
ENSG00000089225.15 | TBX5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TBX5 | hg19_v2_chr12_-_114841703_114841726 | -0.52 | 3.8e-02 | Click! |
Activity profile of TBX5 motif
Sorted Z-values of TBX5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TBX5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_56150910 | 5.04 |
ENST00000424836.2 ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
| chr2_-_175870085 | 4.60 |
ENST00000409156.3 |
CHN1 |
chimerin 1 |
| chr2_-_56150184 | 4.38 |
ENST00000394554.1 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
| chr3_-_120170052 | 3.23 |
ENST00000295633.3 |
FSTL1 |
follistatin-like 1 |
| chr2_-_175869936 | 2.81 |
ENST00000409900.3 |
CHN1 |
chimerin 1 |
| chr11_+_86511569 | 2.44 |
ENST00000441050.1 |
PRSS23 |
protease, serine, 23 |
| chr6_+_121756809 | 2.06 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr21_+_30502806 | 2.01 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chrX_-_34675391 | 1.80 |
ENST00000275954.3 |
TMEM47 |
transmembrane protein 47 |
| chr18_-_52969844 | 1.61 |
ENST00000561831.3 |
TCF4 |
transcription factor 4 |
| chr1_-_153518270 | 1.60 |
ENST00000354332.4 ENST00000368716.4 |
S100A4 |
S100 calcium binding protein A4 |
| chr5_+_82767487 | 1.54 |
ENST00000343200.5 ENST00000342785.4 |
VCAN |
versican |
| chr5_+_82767284 | 1.49 |
ENST00000265077.3 |
VCAN |
versican |
| chr2_-_161350305 | 1.43 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr5_+_82767583 | 1.41 |
ENST00000512590.2 ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN |
versican |
| chr11_+_35198118 | 1.39 |
ENST00000525211.1 ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr4_-_186456766 | 1.38 |
ENST00000284771.6 |
PDLIM3 |
PDZ and LIM domain 3 |
| chr4_-_186456652 | 1.35 |
ENST00000284767.5 ENST00000284770.5 |
PDLIM3 |
PDZ and LIM domain 3 |
| chr6_+_83073952 | 1.31 |
ENST00000543496.1 |
TPBG |
trophoblast glycoprotein |
| chr5_-_54281407 | 1.27 |
ENST00000381403.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr4_-_102268484 | 1.25 |
ENST00000394853.4 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr9_-_75567962 | 1.22 |
ENST00000297785.3 ENST00000376939.1 |
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
| chr5_+_125758813 | 1.16 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr5_+_125758865 | 1.16 |
ENST00000542322.1 ENST00000544396.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr4_-_102268628 | 1.13 |
ENST00000323055.6 ENST00000512215.1 ENST00000394854.3 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr3_+_159570722 | 1.09 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr16_-_67493110 | 1.06 |
ENST00000602876.1 |
ATP6V0D1 |
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr9_-_79307096 | 1.05 |
ENST00000376717.2 ENST00000223609.6 ENST00000443509.2 |
PRUNE2 |
prune homolog 2 (Drosophila) |
| chr17_-_57184260 | 1.05 |
ENST00000376149.3 ENST00000393066.3 |
TRIM37 |
tripartite motif containing 37 |
| chr2_+_5832799 | 1.03 |
ENST00000322002.3 |
SOX11 |
SRY (sex determining region Y)-box 11 |
| chr8_-_18666360 | 1.03 |
ENST00000286485.8 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr17_-_57184064 | 1.02 |
ENST00000262294.7 |
TRIM37 |
tripartite motif containing 37 |
| chr17_-_7297519 | 1.00 |
ENST00000576362.1 ENST00000571078.1 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr11_-_58345569 | 1.00 |
ENST00000528954.1 ENST00000528489.1 |
LPXN |
leupaxin |
| chr17_+_68165657 | 1.00 |
ENST00000243457.3 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr17_-_7297833 | 0.96 |
ENST00000571802.1 ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr1_+_165600083 | 0.95 |
ENST00000367889.3 |
MGST3 |
microsomal glutathione S-transferase 3 |
| chr17_-_15165854 | 0.94 |
ENST00000395936.1 ENST00000395938.2 |
PMP22 |
peripheral myelin protein 22 |
| chr1_-_33168336 | 0.92 |
ENST00000373484.3 |
SYNC |
syncoilin, intermediate filament protein |
| chr1_+_165600436 | 0.91 |
ENST00000367888.4 ENST00000367885.1 ENST00000367884.2 |
MGST3 |
microsomal glutathione S-transferase 3 |
| chr3_-_52002403 | 0.88 |
ENST00000490063.1 ENST00000468324.1 ENST00000497653.1 ENST00000484633.1 |
PCBP4 |
poly(rC) binding protein 4 |
| chr5_+_140864649 | 0.88 |
ENST00000306593.1 |
PCDHGC4 |
protocadherin gamma subfamily C, 4 |
| chr3_+_135741576 | 0.86 |
ENST00000334546.2 |
PPP2R3A |
protein phosphatase 2, regulatory subunit B'', alpha |
| chr22_+_31488433 | 0.85 |
ENST00000455608.1 |
SMTN |
smoothelin |
| chr4_+_78079570 | 0.83 |
ENST00000509972.1 |
CCNG2 |
cyclin G2 |
| chr8_+_32405785 | 0.78 |
ENST00000287842.3 |
NRG1 |
neuregulin 1 |
| chr9_-_130637244 | 0.77 |
ENST00000373156.1 |
AK1 |
adenylate kinase 1 |
| chr11_+_114310102 | 0.75 |
ENST00000265881.5 |
REXO2 |
RNA exonuclease 2 |
| chr5_+_140868717 | 0.74 |
ENST00000252087.1 |
PCDHGC5 |
protocadherin gamma subfamily C, 5 |
| chr8_+_32405728 | 0.74 |
ENST00000523079.1 ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1 |
neuregulin 1 |
| chr6_+_44184653 | 0.72 |
ENST00000573382.2 ENST00000576476.1 |
RP1-302G2.5 |
RP1-302G2.5 |
| chr12_+_1738363 | 0.72 |
ENST00000397196.2 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
| chr11_-_47470591 | 0.71 |
ENST00000524487.1 |
RAPSN |
receptor-associated protein of the synapse |
| chr11_+_114310237 | 0.69 |
ENST00000539119.1 |
REXO2 |
RNA exonuclease 2 |
| chr11_-_47470703 | 0.66 |
ENST00000298854.2 |
RAPSN |
receptor-associated protein of the synapse |
| chr5_+_140810132 | 0.63 |
ENST00000252085.3 |
PCDHGA12 |
protocadherin gamma subfamily A, 12 |
| chr19_+_38755042 | 0.62 |
ENST00000301244.7 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
| chr11_-_47470682 | 0.62 |
ENST00000529341.1 ENST00000352508.3 |
RAPSN |
receptor-associated protein of the synapse |
| chr3_+_132136331 | 0.60 |
ENST00000260818.6 |
DNAJC13 |
DnaJ (Hsp40) homolog, subfamily C, member 13 |
| chr5_-_55290773 | 0.59 |
ENST00000502326.3 ENST00000381298.2 |
IL6ST |
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chr11_+_114310164 | 0.58 |
ENST00000544196.1 ENST00000539754.1 ENST00000539275.1 |
REXO2 |
RNA exonuclease 2 |
| chr6_+_39760129 | 0.57 |
ENST00000274867.4 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chrX_-_106960285 | 0.57 |
ENST00000503515.1 ENST00000372397.2 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr5_-_54281491 | 0.57 |
ENST00000381405.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chrX_-_134049262 | 0.56 |
ENST00000370783.3 |
MOSPD1 |
motile sperm domain containing 1 |
| chr1_+_101702417 | 0.56 |
ENST00000305352.6 |
S1PR1 |
sphingosine-1-phosphate receptor 1 |
| chr7_+_40174565 | 0.55 |
ENST00000309930.5 ENST00000401647.2 ENST00000335693.4 ENST00000413931.1 ENST00000416370.1 ENST00000540834.1 |
C7orf10 |
succinylCoA:glutarate-CoA transferase |
| chr12_+_81101277 | 0.55 |
ENST00000228641.3 |
MYF6 |
myogenic factor 6 (herculin) |
| chr12_-_102455902 | 0.53 |
ENST00000240079.6 |
CCDC53 |
coiled-coil domain containing 53 |
| chr12_-_102455846 | 0.53 |
ENST00000545679.1 |
CCDC53 |
coiled-coil domain containing 53 |
| chrX_+_95939638 | 0.53 |
ENST00000373061.3 ENST00000373054.4 ENST00000355827.4 |
DIAPH2 |
diaphanous-related formin 2 |
| chr14_-_82000140 | 0.53 |
ENST00000555824.1 ENST00000557372.1 ENST00000336735.4 |
SEL1L |
sel-1 suppressor of lin-12-like (C. elegans) |
| chr5_-_16738451 | 0.51 |
ENST00000274203.9 ENST00000515803.1 |
MYO10 |
myosin X |
| chr19_+_38755203 | 0.51 |
ENST00000587090.1 ENST00000454580.3 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
| chr18_+_33877654 | 0.51 |
ENST00000257209.4 ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr2_+_234668894 | 0.50 |
ENST00000305208.5 ENST00000608383.1 ENST00000360418.3 |
UGT1A8 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr1_+_19923454 | 0.50 |
ENST00000602662.1 ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1 MINOS1 |
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr14_-_24664540 | 0.49 |
ENST00000530563.1 ENST00000528895.1 ENST00000528669.1 ENST00000532632.1 |
TM9SF1 |
transmembrane 9 superfamily member 1 |
| chr11_+_1940786 | 0.48 |
ENST00000278317.6 ENST00000381561.4 ENST00000381548.3 ENST00000360603.3 ENST00000381549.3 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
| chr17_+_47572647 | 0.48 |
ENST00000172229.3 |
NGFR |
nerve growth factor receptor |
| chr16_-_53537105 | 0.48 |
ENST00000568596.1 ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP |
AKT interacting protein |
| chr7_-_137028534 | 0.47 |
ENST00000348225.2 |
PTN |
pleiotrophin |
| chr17_+_58755184 | 0.46 |
ENST00000589222.1 ENST00000407086.3 ENST00000390652.5 |
BCAS3 |
breast carcinoma amplified sequence 3 |
| chr4_-_120133661 | 0.45 |
ENST00000503243.1 ENST00000326780.3 |
RP11-455G16.1 |
Uncharacterized protein |
| chr7_-_137028498 | 0.45 |
ENST00000393083.2 |
PTN |
pleiotrophin |
| chr7_-_27135591 | 0.44 |
ENST00000343060.4 ENST00000355633.5 |
HOXA1 |
homeobox A1 |
| chr11_-_107590383 | 0.44 |
ENST00000525934.1 ENST00000531293.1 |
SLN |
sarcolipin |
| chr12_+_6833437 | 0.43 |
ENST00000534947.1 ENST00000541866.1 ENST00000534877.1 ENST00000538753.1 |
COPS7A |
COP9 signalosome subunit 7A |
| chr11_-_85779971 | 0.43 |
ENST00000393346.3 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
| chr1_-_205290865 | 0.43 |
ENST00000367157.3 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
| chr11_-_72385437 | 0.42 |
ENST00000418754.2 ENST00000542969.2 ENST00000334456.5 |
PDE2A |
phosphodiesterase 2A, cGMP-stimulated |
| chr6_+_39760783 | 0.42 |
ENST00000398904.2 ENST00000538976.1 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr7_-_76255444 | 0.42 |
ENST00000454397.1 |
POMZP3 |
POM121 and ZP3 fusion |
| chr14_+_90864504 | 0.41 |
ENST00000544280.2 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
| chr12_-_49351228 | 0.41 |
ENST00000541959.1 ENST00000447318.2 |
ARF3 |
ADP-ribosylation factor 3 |
| chr7_+_107220660 | 0.40 |
ENST00000465919.1 ENST00000445771.2 ENST00000479917.1 ENST00000421217.1 ENST00000457837.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr17_-_26220366 | 0.40 |
ENST00000460380.2 ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9 RP1-66C13.4 |
LYR motif containing 9 Uncharacterized protein |
| chr2_-_191878874 | 0.39 |
ENST00000392322.3 ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1 |
signal transducer and activator of transcription 1, 91kDa |
| chr2_+_85766280 | 0.38 |
ENST00000306434.3 |
MAT2A |
methionine adenosyltransferase II, alpha |
| chr11_+_1940925 | 0.37 |
ENST00000453458.1 ENST00000381557.2 ENST00000381589.3 ENST00000381579.3 ENST00000381563.4 ENST00000344578.4 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
| chr2_-_9770706 | 0.37 |
ENST00000381844.4 |
YWHAQ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr11_-_2182388 | 0.37 |
ENST00000421783.1 ENST00000397262.1 ENST00000250971.3 ENST00000381330.4 ENST00000397270.1 |
INS INS-IGF2 |
insulin INS-IGF2 readthrough |
| chr2_+_234601512 | 0.36 |
ENST00000305139.6 |
UGT1A6 |
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr4_+_90823130 | 0.35 |
ENST00000508372.1 |
MMRN1 |
multimerin 1 |
| chr1_-_241520525 | 0.35 |
ENST00000366565.1 |
RGS7 |
regulator of G-protein signaling 7 |
| chr2_+_201676908 | 0.35 |
ENST00000409226.1 ENST00000452790.2 |
BZW1 |
basic leucine zipper and W2 domains 1 |
| chrX_+_95939711 | 0.34 |
ENST00000373049.4 ENST00000324765.8 |
DIAPH2 |
diaphanous-related formin 2 |
| chr16_-_67190152 | 0.34 |
ENST00000486556.1 |
TRADD |
TNFRSF1A-associated via death domain |
| chr2_+_29336855 | 0.33 |
ENST00000404424.1 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
| chr13_-_36429763 | 0.33 |
ENST00000379893.1 |
DCLK1 |
doublecortin-like kinase 1 |
| chr1_+_13910194 | 0.33 |
ENST00000376057.4 ENST00000510906.1 |
PDPN |
podoplanin |
| chr22_+_38597889 | 0.32 |
ENST00000338483.2 ENST00000538320.1 ENST00000538999.1 ENST00000441709.1 |
MAFF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr17_+_35849937 | 0.32 |
ENST00000394389.4 |
DUSP14 |
dual specificity phosphatase 14 |
| chr21_+_38445539 | 0.31 |
ENST00000418766.1 ENST00000450533.1 ENST00000438055.1 ENST00000355666.1 ENST00000540756.1 ENST00000399010.1 |
TTC3 |
tetratricopeptide repeat domain 3 |
| chr6_-_33239612 | 0.31 |
ENST00000482399.1 ENST00000445902.2 |
VPS52 |
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr1_-_144995002 | 0.31 |
ENST00000369356.4 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr13_+_115047097 | 0.30 |
ENST00000351487.5 |
UPF3A |
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr3_+_35722487 | 0.30 |
ENST00000441454.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr5_+_138089100 | 0.30 |
ENST00000520339.1 ENST00000355078.5 ENST00000302763.7 ENST00000518910.1 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr22_+_38609538 | 0.29 |
ENST00000407965.1 |
MAFF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr9_+_125133315 | 0.29 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr3_+_133292759 | 0.28 |
ENST00000431519.2 |
CDV3 |
CDV3 homolog (mouse) |
| chr14_-_103523745 | 0.28 |
ENST00000361246.2 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
| chr19_-_45826125 | 0.28 |
ENST00000221476.3 |
CKM |
creatine kinase, muscle |
| chr1_+_13910479 | 0.28 |
ENST00000509009.1 |
PDPN |
podoplanin |
| chr14_+_21467414 | 0.28 |
ENST00000554422.1 ENST00000298681.4 |
SLC39A2 |
solute carrier family 39 (zinc transporter), member 2 |
| chr12_+_53645870 | 0.28 |
ENST00000329548.4 |
MFSD5 |
major facilitator superfamily domain containing 5 |
| chr2_+_64068074 | 0.27 |
ENST00000394417.2 ENST00000484142.1 ENST00000482668.1 ENST00000467648.2 |
UGP2 |
UDP-glucose pyrophosphorylase 2 |
| chr2_+_201676256 | 0.27 |
ENST00000452206.1 ENST00000410110.2 ENST00000409600.1 |
BZW1 |
basic leucine zipper and W2 domains 1 |
| chr2_-_152589670 | 0.27 |
ENST00000604864.1 ENST00000603639.1 |
NEB |
nebulin |
| chrX_-_106959631 | 0.27 |
ENST00000486554.1 ENST00000372390.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr7_-_130080818 | 0.27 |
ENST00000343969.5 ENST00000541543.1 ENST00000489512.1 |
CEP41 |
centrosomal protein 41kDa |
| chr9_-_34637718 | 0.26 |
ENST00000378892.1 ENST00000277010.4 |
SIGMAR1 |
sigma non-opioid intracellular receptor 1 |
| chr7_+_86975001 | 0.26 |
ENST00000412227.2 ENST00000331536.3 |
CROT |
carnitine O-octanoyltransferase |
| chr1_+_202995611 | 0.26 |
ENST00000367240.2 |
PPFIA4 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr12_+_12764773 | 0.26 |
ENST00000228865.2 |
CREBL2 |
cAMP responsive element binding protein-like 2 |
| chr14_+_78870030 | 0.26 |
ENST00000553631.1 ENST00000554719.1 |
NRXN3 |
neurexin 3 |
| chr12_+_8234807 | 0.26 |
ENST00000339754.5 |
NECAP1 |
NECAP endocytosis associated 1 |
| chr6_+_30035307 | 0.25 |
ENST00000376765.2 ENST00000376763.1 |
PPP1R11 |
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr15_+_90808919 | 0.25 |
ENST00000379095.3 |
NGRN |
neugrin, neurite outgrowth associated |
| chr22_-_31688431 | 0.25 |
ENST00000402249.3 ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
| chr1_+_13910757 | 0.25 |
ENST00000376061.4 ENST00000513143.1 |
PDPN |
podoplanin |
| chrX_+_153672468 | 0.25 |
ENST00000393600.3 |
FAM50A |
family with sequence similarity 50, member A |
| chr14_+_61995722 | 0.24 |
ENST00000556347.1 |
RP11-47I22.4 |
RP11-47I22.4 |
| chr22_-_38699003 | 0.24 |
ENST00000451964.1 |
CSNK1E |
casein kinase 1, epsilon |
| chr12_-_52887034 | 0.24 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chr6_+_30034966 | 0.23 |
ENST00000376769.2 |
PPP1R11 |
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr3_-_18466026 | 0.23 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr16_+_66400533 | 0.23 |
ENST00000341529.3 |
CDH5 |
cadherin 5, type 2 (vascular endothelium) |
| chr7_+_30951461 | 0.22 |
ENST00000311813.4 |
AQP1 |
aquaporin 1 (Colton blood group) |
| chr15_-_41099648 | 0.22 |
ENST00000220496.4 |
DNAJC17 |
DnaJ (Hsp40) homolog, subfamily C, member 17 |
| chr8_-_99954788 | 0.21 |
ENST00000523601.1 |
STK3 |
serine/threonine kinase 3 |
| chr18_+_3449695 | 0.21 |
ENST00000343820.5 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr19_+_50380917 | 0.21 |
ENST00000535102.2 |
TBC1D17 |
TBC1 domain family, member 17 |
| chr14_-_68283291 | 0.21 |
ENST00000555452.1 ENST00000347230.4 |
ZFYVE26 |
zinc finger, FYVE domain containing 26 |
| chr14_-_107211459 | 0.21 |
ENST00000390636.2 |
IGHV3-73 |
immunoglobulin heavy variable 3-73 |
| chr5_-_180229833 | 0.20 |
ENST00000307826.4 |
MGAT1 |
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr5_-_140070897 | 0.20 |
ENST00000448240.1 ENST00000438307.2 ENST00000415192.2 ENST00000457527.2 ENST00000307633.3 ENST00000507746.1 ENST00000431330.2 |
HARS |
histidyl-tRNA synthetase |
| chr6_-_33239712 | 0.20 |
ENST00000436044.2 |
VPS52 |
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr12_-_49351303 | 0.20 |
ENST00000256682.4 |
ARF3 |
ADP-ribosylation factor 3 |
| chrX_-_109561294 | 0.20 |
ENST00000372059.2 ENST00000262844.5 |
AMMECR1 |
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr11_+_18343800 | 0.20 |
ENST00000453096.2 |
GTF2H1 |
general transcription factor IIH, polypeptide 1, 62kDa |
| chr7_+_100450328 | 0.20 |
ENST00000540482.1 ENST00000418037.1 ENST00000428758.1 ENST00000275729.3 ENST00000415287.1 ENST00000354161.3 ENST00000416675.1 |
SLC12A9 |
solute carrier family 12, member 9 |
| chr9_+_125137565 | 0.19 |
ENST00000373698.5 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chrX_+_135618258 | 0.18 |
ENST00000440515.1 ENST00000456412.1 |
VGLL1 |
vestigial like 1 (Drosophila) |
| chr11_+_18344106 | 0.18 |
ENST00000534641.1 ENST00000525831.1 ENST00000265963.4 |
GTF2H1 |
general transcription factor IIH, polypeptide 1, 62kDa |
| chr9_-_6015607 | 0.17 |
ENST00000259569.5 |
RANBP6 |
RAN binding protein 6 |
| chr17_+_27055798 | 0.17 |
ENST00000268766.6 |
NEK8 |
NIMA-related kinase 8 |
| chr11_+_34073269 | 0.17 |
ENST00000389645.3 |
CAPRIN1 |
cell cycle associated protein 1 |
| chrX_+_21392553 | 0.17 |
ENST00000279451.4 |
CNKSR2 |
connector enhancer of kinase suppressor of Ras 2 |
| chr1_+_35734562 | 0.16 |
ENST00000314607.6 ENST00000373297.2 |
ZMYM4 |
zinc finger, MYM-type 4 |
| chr19_-_46526304 | 0.16 |
ENST00000008938.4 |
PGLYRP1 |
peptidoglycan recognition protein 1 |
| chr2_-_219925189 | 0.16 |
ENST00000295731.6 |
IHH |
indian hedgehog |
| chr16_+_2546033 | 0.16 |
ENST00000564543.1 ENST00000434757.2 |
RP11-20I23.1 TBC1D24 |
Uncharacterized protein TBC1 domain family, member 24 |
| chr14_-_37051798 | 0.16 |
ENST00000258829.5 |
NKX2-8 |
NK2 homeobox 8 |
| chr14_-_45603657 | 0.16 |
ENST00000396062.3 |
FKBP3 |
FK506 binding protein 3, 25kDa |
| chr13_-_46543805 | 0.15 |
ENST00000378921.2 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
| chr10_+_135340859 | 0.15 |
ENST00000252945.3 ENST00000421586.1 ENST00000418356.1 |
CYP2E1 |
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr14_-_107114267 | 0.15 |
ENST00000454421.2 |
IGHV3-64 |
immunoglobulin heavy variable 3-64 |
| chr11_+_36589547 | 0.15 |
ENST00000299440.5 |
RAG1 |
recombination activating gene 1 |
| chr11_-_34535297 | 0.14 |
ENST00000532417.1 |
ELF5 |
E74-like factor 5 (ets domain transcription factor) |
| chr19_-_49622348 | 0.14 |
ENST00000408991.2 |
C19orf73 |
chromosome 19 open reading frame 73 |
| chr10_+_76586348 | 0.14 |
ENST00000372724.1 ENST00000287239.4 ENST00000372714.1 |
KAT6B |
K(lysine) acetyltransferase 6B |
| chr14_-_23904861 | 0.14 |
ENST00000355349.3 |
MYH7 |
myosin, heavy chain 7, cardiac muscle, beta |
| chr19_+_41107249 | 0.14 |
ENST00000396819.3 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr11_-_45939565 | 0.13 |
ENST00000525192.1 ENST00000378750.5 |
PEX16 |
peroxisomal biogenesis factor 16 |
| chr18_+_3449821 | 0.13 |
ENST00000407501.2 ENST00000405385.3 ENST00000546979.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr1_+_15736359 | 0.13 |
ENST00000375980.4 |
EFHD2 |
EF-hand domain family, member D2 |
| chr1_+_16062820 | 0.13 |
ENST00000294454.5 |
SLC25A34 |
solute carrier family 25, member 34 |
| chr6_-_31080336 | 0.12 |
ENST00000259870.3 |
C6orf15 |
chromosome 6 open reading frame 15 |
| chr7_-_86974785 | 0.12 |
ENST00000432193.1 ENST00000421293.1 ENST00000542586.1 ENST00000359941.5 ENST00000416560.1 |
TP53TG1 |
TP53 target 1 (non-protein coding) |
| chr19_+_50380682 | 0.12 |
ENST00000221543.5 |
TBC1D17 |
TBC1 domain family, member 17 |
| chr16_+_3162557 | 0.12 |
ENST00000382192.3 ENST00000219091.4 ENST00000444510.2 ENST00000414351.1 |
ZNF205 |
zinc finger protein 205 |
| chr7_-_72972319 | 0.11 |
ENST00000223368.2 |
BCL7B |
B-cell CLL/lymphoma 7B |
| chr8_+_26149007 | 0.11 |
ENST00000380737.3 ENST00000524169.1 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
| chr1_+_184356188 | 0.11 |
ENST00000235307.6 |
C1orf21 |
chromosome 1 open reading frame 21 |
| chrX_-_53449593 | 0.11 |
ENST00000375340.6 ENST00000322213.4 |
SMC1A |
structural maintenance of chromosomes 1A |
| chr11_-_45939374 | 0.11 |
ENST00000533151.1 ENST00000241041.3 |
PEX16 |
peroxisomal biogenesis factor 16 |
| chr10_-_102279586 | 0.11 |
ENST00000370345.3 ENST00000451524.1 ENST00000370329.5 |
SEC31B |
SEC31 homolog B (S. cerevisiae) |
| chr19_+_55085248 | 0.11 |
ENST00000391738.3 ENST00000251376.3 ENST00000391737.1 ENST00000396321.2 ENST00000418536.2 ENST00000448689.1 |
LILRA2 LILRB1 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 2 leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 9.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.6 | 1.8 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.4 | 2.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.2 | 1.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.2 | 2.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 0.9 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.2 | 0.9 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.2 | 6.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 0.6 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) oncostatin-M receptor activity(GO:0004924) interleukin-6 binding(GO:0019981) |
| 0.2 | 1.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 7.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 0.5 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 1.6 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.5 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 2.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.8 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.3 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 1.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.4 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.2 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 2.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 2.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.5 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.1 | 0.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 1.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 3.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.6 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.0 | 0.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 1.9 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 1.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.1 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 1.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 3.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.8 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 2.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 2.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 6.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 1.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.2 | 1.9 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.6 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 2.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.5 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.5 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.6 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.9 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 1.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.2 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 1.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 5.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.9 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 1.0 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 5.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 8.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 3.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0098793 | presynapse(GO:0098793) |
| 0.0 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0072589 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 1.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 9.4 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.8 | 2.4 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.7 | 2.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.7 | 2.1 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.7 | 2.0 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.4 | 1.2 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.3 | 1.0 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.3 | 0.9 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.3 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.3 | 0.9 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.3 | 1.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.3 | 1.8 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.3 | 1.0 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 1.5 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.2 | 7.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 0.6 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.2 | 4.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.2 | 1.4 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.2 | 0.5 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.2 | 0.6 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.1 | 0.6 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.4 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.1 | 0.4 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.1 | 0.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 0.4 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.1 | 0.4 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.1 | 0.3 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.4 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 0.8 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.5 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 0.4 | GO:2000697 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.1 | 0.6 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.5 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 0.1 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.1 | 0.2 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.1 | 0.2 | GO:0072229 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.2 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 0.3 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.4 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 1.8 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.1 | 1.0 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 1.1 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.9 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.7 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.4 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.2 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.3 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.2 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0072423 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.1 | GO:0035548 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) |
| 0.0 | 0.4 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 1.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 1.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.5 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0007418 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.8 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.2 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 3.7 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0001712 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.5 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 2.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.2 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.5 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 7.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.4 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 2.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 9.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.9 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |