Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
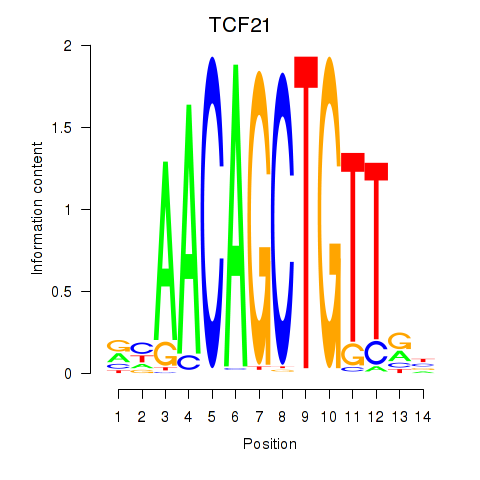
Results for TCF21
Z-value: 0.75
Transcription factors associated with TCF21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF21
|
ENSG00000118526.6 | TCF21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF21 | hg19_v2_chr6_+_134210243_134210276 | -0.01 | 9.6e-01 | Click! |
Activity profile of TCF21 motif
Sorted Z-values of TCF21 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF21
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_31230650 | 2.18 |
ENST00000294507.3 |
LAPTM5 |
lysosomal protein transmembrane 5 |
| chr2_-_158345462 | 2.00 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr1_-_160681593 | 1.65 |
ENST00000368045.3 ENST00000368046.3 |
CD48 |
CD48 molecule |
| chr5_-_95158644 | 1.29 |
ENST00000237858.6 |
GLRX |
glutaredoxin (thioltransferase) |
| chr6_+_13182751 | 1.27 |
ENST00000415087.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr11_+_1874200 | 1.23 |
ENST00000311604.3 |
LSP1 |
lymphocyte-specific protein 1 |
| chr6_+_12958137 | 1.15 |
ENST00000457702.2 ENST00000379345.2 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr1_-_173176452 | 1.12 |
ENST00000281834.3 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr3_+_8775466 | 1.08 |
ENST00000343849.2 ENST00000397368.2 |
CAV3 |
caveolin 3 |
| chr14_-_23288930 | 1.06 |
ENST00000554517.1 ENST00000285850.7 ENST00000397529.2 ENST00000555702.1 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr2_-_225811747 | 1.05 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr2_+_33701286 | 1.04 |
ENST00000403687.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr17_+_45286387 | 1.03 |
ENST00000572316.1 ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr22_+_23264766 | 1.01 |
ENST00000390331.2 |
IGLC7 |
immunoglobulin lambda constant 7 |
| chr1_-_9129735 | 0.95 |
ENST00000377424.4 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr3_-_182833863 | 0.87 |
ENST00000492597.1 |
MCCC1 |
methylcrotonoyl-CoA carboxylase 1 (alpha) |
| chr5_+_95066823 | 0.86 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr15_+_83776137 | 0.84 |
ENST00000322019.9 |
TM6SF1 |
transmembrane 6 superfamily member 1 |
| chr1_+_111772314 | 0.82 |
ENST00000466741.1 ENST00000477185.2 |
CHI3L2 |
chitinase 3-like 2 |
| chr1_-_9129598 | 0.81 |
ENST00000535586.1 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr16_+_30383613 | 0.76 |
ENST00000568749.1 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr11_-_615570 | 0.75 |
ENST00000525445.1 ENST00000348655.6 ENST00000397566.1 |
IRF7 |
interferon regulatory factor 7 |
| chr9_+_2029019 | 0.74 |
ENST00000382194.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr17_+_45286706 | 0.72 |
ENST00000393450.1 ENST00000572303.1 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr1_+_79115503 | 0.72 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr1_-_9129895 | 0.71 |
ENST00000473209.1 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr8_+_38585704 | 0.71 |
ENST00000519416.1 ENST00000520615.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr10_+_35415719 | 0.69 |
ENST00000474362.1 ENST00000374721.3 |
CREM |
cAMP responsive element modulator |
| chr2_-_175629135 | 0.68 |
ENST00000409542.1 ENST00000409219.1 |
CHRNA1 |
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr11_+_7597639 | 0.65 |
ENST00000533792.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr1_-_9129631 | 0.64 |
ENST00000377414.3 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr5_+_49963239 | 0.62 |
ENST00000505554.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr22_-_23922410 | 0.62 |
ENST00000249053.3 |
IGLL1 |
immunoglobulin lambda-like polypeptide 1 |
| chr7_+_150434430 | 0.59 |
ENST00000358647.3 |
GIMAP5 |
GTPase, IMAP family member 5 |
| chrX_-_106960285 | 0.57 |
ENST00000503515.1 ENST00000372397.2 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr3_-_105588231 | 0.57 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chrX_-_11445856 | 0.55 |
ENST00000380736.1 |
ARHGAP6 |
Rho GTPase activating protein 6 |
| chr3_-_81792780 | 0.54 |
ENST00000489715.1 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
| chr12_+_104458235 | 0.53 |
ENST00000229330.4 |
HCFC2 |
host cell factor C2 |
| chr5_+_49962772 | 0.51 |
ENST00000281631.5 ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chrX_-_78622805 | 0.50 |
ENST00000373298.2 |
ITM2A |
integral membrane protein 2A |
| chr2_-_152590946 | 0.49 |
ENST00000172853.10 |
NEB |
nebulin |
| chr7_+_142498725 | 0.47 |
ENST00000466254.1 |
TRBC2 |
T cell receptor beta constant 2 |
| chr6_-_31697255 | 0.47 |
ENST00000436437.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr11_+_58910295 | 0.45 |
ENST00000420244.1 |
FAM111A |
family with sequence similarity 111, member A |
| chr2_+_231280908 | 0.44 |
ENST00000427101.2 ENST00000432979.1 |
SP100 |
SP100 nuclear antigen |
| chr16_+_56970567 | 0.43 |
ENST00000563911.1 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr6_+_44184653 | 0.43 |
ENST00000573382.2 ENST00000576476.1 |
RP1-302G2.5 |
RP1-302G2.5 |
| chr1_+_212738676 | 0.41 |
ENST00000366981.4 ENST00000366987.2 |
ATF3 |
activating transcription factor 3 |
| chr11_+_1944054 | 0.41 |
ENST00000397301.1 ENST00000397304.2 ENST00000446240.1 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
| chr11_+_32112431 | 0.40 |
ENST00000054950.3 |
RCN1 |
reticulocalbin 1, EF-hand calcium binding domain |
| chr3_+_179065474 | 0.40 |
ENST00000471841.1 ENST00000280653.7 |
MFN1 |
mitofusin 1 |
| chr13_+_51483814 | 0.40 |
ENST00000336617.3 ENST00000422660.1 |
RNASEH2B |
ribonuclease H2, subunit B |
| chr11_+_58910201 | 0.40 |
ENST00000528737.1 |
FAM111A |
family with sequence similarity 111, member A |
| chr7_-_37024665 | 0.39 |
ENST00000396040.2 |
ELMO1 |
engulfment and cell motility 1 |
| chr4_-_4543700 | 0.39 |
ENST00000505286.1 ENST00000306200.2 |
STX18 |
syntaxin 18 |
| chr10_+_35415978 | 0.38 |
ENST00000429130.3 ENST00000469949.2 ENST00000460270.1 |
CREM |
cAMP responsive element modulator |
| chr2_-_214016314 | 0.38 |
ENST00000434687.1 ENST00000374319.4 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
| chr6_-_26250835 | 0.37 |
ENST00000446824.2 |
HIST1H3F |
histone cluster 1, H3f |
| chr11_-_615942 | 0.37 |
ENST00000397562.3 ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7 |
interferon regulatory factor 7 |
| chr19_-_45826125 | 0.37 |
ENST00000221476.3 |
CKM |
creatine kinase, muscle |
| chr3_+_178525080 | 0.37 |
ENST00000358316.3 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr1_-_39339777 | 0.36 |
ENST00000397572.2 |
MYCBP |
MYC binding protein |
| chr6_-_31697563 | 0.36 |
ENST00000375789.2 ENST00000416410.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr12_+_112563335 | 0.36 |
ENST00000549358.1 ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1 |
TRAF-type zinc finger domain containing 1 |
| chr19_+_18208603 | 0.36 |
ENST00000262811.6 |
MAST3 |
microtubule associated serine/threonine kinase 3 |
| chr11_-_82997371 | 0.36 |
ENST00000525503.1 |
CCDC90B |
coiled-coil domain containing 90B |
| chr5_+_170288856 | 0.35 |
ENST00000523189.1 |
RANBP17 |
RAN binding protein 17 |
| chr11_+_33061543 | 0.34 |
ENST00000432887.1 ENST00000528898.1 ENST00000531632.2 |
TCP11L1 |
t-complex 11, testis-specific-like 1 |
| chr17_+_80416482 | 0.34 |
ENST00000309794.11 ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF |
nuclear prelamin A recognition factor |
| chrX_+_48432892 | 0.33 |
ENST00000376759.3 ENST00000430348.2 |
RBM3 |
RNA binding motif (RNP1, RRM) protein 3 |
| chr11_+_125496400 | 0.33 |
ENST00000524737.1 |
CHEK1 |
checkpoint kinase 1 |
| chrX_+_148622138 | 0.32 |
ENST00000450602.2 ENST00000441248.1 |
CXorf40A |
chromosome X open reading frame 40A |
| chr2_-_175629164 | 0.32 |
ENST00000409323.1 ENST00000261007.5 ENST00000348749.5 |
CHRNA1 |
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr17_+_68165657 | 0.31 |
ENST00000243457.3 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr16_-_31076332 | 0.31 |
ENST00000539836.3 ENST00000535577.1 ENST00000442862.2 |
ZNF668 |
zinc finger protein 668 |
| chr2_+_231280954 | 0.30 |
ENST00000409824.1 ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100 |
SP100 nuclear antigen |
| chr11_-_82997420 | 0.30 |
ENST00000455220.2 ENST00000529689.1 |
CCDC90B |
coiled-coil domain containing 90B |
| chr15_+_63354769 | 0.29 |
ENST00000558910.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr11_+_125496619 | 0.29 |
ENST00000532669.1 ENST00000278916.3 |
CHEK1 |
checkpoint kinase 1 |
| chr2_-_128400788 | 0.29 |
ENST00000409286.1 |
LIMS2 |
LIM and senescent cell antigen-like domains 2 |
| chr11_-_117103208 | 0.29 |
ENST00000320934.3 ENST00000530269.1 |
PCSK7 |
proprotein convertase subtilisin/kexin type 7 |
| chr17_+_71229346 | 0.29 |
ENST00000535032.2 ENST00000582793.1 |
C17orf80 |
chromosome 17 open reading frame 80 |
| chr4_-_46911248 | 0.28 |
ENST00000355591.3 ENST00000505102.1 |
COX7B2 |
cytochrome c oxidase subunit VIIb2 |
| chr4_+_146403912 | 0.28 |
ENST00000507367.1 ENST00000394092.2 ENST00000515385.1 |
SMAD1 |
SMAD family member 1 |
| chr10_-_79398127 | 0.28 |
ENST00000372443.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr11_+_111957497 | 0.28 |
ENST00000375549.3 ENST00000528182.1 ENST00000528048.1 ENST00000528021.1 ENST00000526592.1 ENST00000525291.1 |
SDHD |
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr16_-_46655538 | 0.27 |
ENST00000303383.3 |
SHCBP1 |
SHC SH2-domain binding protein 1 |
| chr20_-_52210368 | 0.27 |
ENST00000371471.2 |
ZNF217 |
zinc finger protein 217 |
| chr6_+_31554962 | 0.27 |
ENST00000376092.3 ENST00000376086.3 ENST00000303757.8 ENST00000376093.2 ENST00000376102.3 |
LST1 |
leukocyte specific transcript 1 |
| chr17_-_33864772 | 0.27 |
ENST00000361112.4 |
SLFN12L |
schlafen family member 12-like |
| chr1_-_161993422 | 0.26 |
ENST00000367940.2 |
OLFML2B |
olfactomedin-like 2B |
| chr11_+_125496124 | 0.26 |
ENST00000533778.2 ENST00000534070.1 |
CHEK1 |
checkpoint kinase 1 |
| chr9_-_89562104 | 0.26 |
ENST00000298743.7 |
GAS1 |
growth arrest-specific 1 |
| chr6_+_31555045 | 0.26 |
ENST00000396101.3 ENST00000490742.1 |
LST1 |
leukocyte specific transcript 1 |
| chr22_+_38071615 | 0.26 |
ENST00000215909.5 |
LGALS1 |
lectin, galactoside-binding, soluble, 1 |
| chr12_+_72233487 | 0.26 |
ENST00000482439.2 ENST00000550746.1 ENST00000491063.1 ENST00000319106.8 ENST00000485960.2 ENST00000393309.3 |
TBC1D15 |
TBC1 domain family, member 15 |
| chr4_-_46911223 | 0.26 |
ENST00000396533.1 |
COX7B2 |
cytochrome c oxidase subunit VIIb2 |
| chr22_-_23922448 | 0.25 |
ENST00000438703.1 ENST00000330377.2 |
IGLL1 |
immunoglobulin lambda-like polypeptide 1 |
| chr4_-_186347099 | 0.25 |
ENST00000505357.1 ENST00000264689.6 |
UFSP2 |
UFM1-specific peptidase 2 |
| chr1_-_230513367 | 0.25 |
ENST00000321327.2 ENST00000525115.1 |
PGBD5 |
piggyBac transposable element derived 5 |
| chr3_+_197677379 | 0.24 |
ENST00000442341.1 |
RPL35A |
ribosomal protein L35a |
| chr12_-_104443890 | 0.24 |
ENST00000547583.1 ENST00000360814.4 ENST00000546851.1 |
GLT8D2 |
glycosyltransferase 8 domain containing 2 |
| chr7_-_99698338 | 0.24 |
ENST00000354230.3 ENST00000425308.1 |
MCM7 |
minichromosome maintenance complex component 7 |
| chr22_+_40742512 | 0.23 |
ENST00000454266.2 ENST00000342312.6 |
ADSL |
adenylosuccinate lyase |
| chr1_+_14075865 | 0.23 |
ENST00000413440.1 |
PRDM2 |
PR domain containing 2, with ZNF domain |
| chr11_+_74660278 | 0.23 |
ENST00000263672.6 ENST00000530257.1 ENST00000526361.1 ENST00000532972.1 |
SPCS2 |
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr10_-_79398250 | 0.23 |
ENST00000286627.5 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr5_-_179050066 | 0.22 |
ENST00000329433.6 ENST00000510411.1 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr3_+_141106643 | 0.22 |
ENST00000514251.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr17_+_71228740 | 0.22 |
ENST00000268942.8 ENST00000359042.2 |
C17orf80 |
chromosome 17 open reading frame 80 |
| chr2_+_44001172 | 0.22 |
ENST00000260605.8 ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1 |
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr11_-_130786400 | 0.22 |
ENST00000265909.4 |
SNX19 |
sorting nexin 19 |
| chrX_+_148622513 | 0.22 |
ENST00000393985.3 ENST00000423421.1 ENST00000423540.2 ENST00000434353.2 ENST00000514208.1 |
CXorf40A |
chromosome X open reading frame 40A |
| chr15_+_63796779 | 0.22 |
ENST00000561442.1 ENST00000560070.1 ENST00000540797.1 ENST00000380324.3 ENST00000268049.7 ENST00000536001.1 ENST00000539772.1 |
USP3 |
ubiquitin specific peptidase 3 |
| chr2_-_228028829 | 0.21 |
ENST00000396625.3 ENST00000329662.7 |
COL4A4 |
collagen, type IV, alpha 4 |
| chr19_+_1269324 | 0.21 |
ENST00000589710.1 ENST00000588230.1 ENST00000413636.2 ENST00000586472.1 ENST00000589686.1 ENST00000444172.2 ENST00000587323.1 ENST00000320936.5 ENST00000587896.1 ENST00000589235.1 ENST00000591659.1 |
CIRBP |
cold inducible RNA binding protein |
| chr11_+_64002292 | 0.21 |
ENST00000426086.2 |
VEGFB |
vascular endothelial growth factor B |
| chr15_+_63340775 | 0.20 |
ENST00000559281.1 ENST00000317516.7 |
TPM1 |
tropomyosin 1 (alpha) |
| chr15_+_57998821 | 0.20 |
ENST00000299638.3 |
POLR2M |
polymerase (RNA) II (DNA directed) polypeptide M |
| chr11_-_82997013 | 0.20 |
ENST00000529073.1 ENST00000529611.1 |
CCDC90B |
coiled-coil domain containing 90B |
| chr2_-_191115229 | 0.20 |
ENST00000409820.2 ENST00000410045.1 |
HIBCH |
3-hydroxyisobutyryl-CoA hydrolase |
| chr17_+_71228793 | 0.20 |
ENST00000426147.2 |
C17orf80 |
chromosome 17 open reading frame 80 |
| chr2_+_97202480 | 0.20 |
ENST00000357485.3 |
ARID5A |
AT rich interactive domain 5A (MRF1-like) |
| chr8_-_99837856 | 0.20 |
ENST00000518165.1 ENST00000419617.2 |
STK3 |
serine/threonine kinase 3 |
| chr6_-_39197226 | 0.19 |
ENST00000359534.3 |
KCNK5 |
potassium channel, subfamily K, member 5 |
| chr2_+_79412357 | 0.19 |
ENST00000466387.1 |
CTNNA2 |
catenin (cadherin-associated protein), alpha 2 |
| chr7_-_132766800 | 0.19 |
ENST00000542753.1 ENST00000448878.1 |
CHCHD3 |
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr12_-_6960407 | 0.19 |
ENST00000540683.1 ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3 |
cell division cycle associated 3 |
| chrX_-_107334750 | 0.19 |
ENST00000340200.5 ENST00000372296.1 ENST00000372295.1 ENST00000361815.5 |
PSMD10 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr17_+_16593539 | 0.19 |
ENST00000340621.5 ENST00000399273.1 ENST00000443444.2 ENST00000360524.8 ENST00000456009.1 |
CCDC144A |
coiled-coil domain containing 144A |
| chr19_-_4867665 | 0.19 |
ENST00000592528.1 ENST00000589494.1 ENST00000585479.1 ENST00000221957.4 |
PLIN3 |
perilipin 3 |
| chr11_-_111957451 | 0.19 |
ENST00000504148.2 ENST00000541231.1 |
TIMM8B |
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr7_-_132766818 | 0.18 |
ENST00000262570.5 |
CHCHD3 |
coiled-coil-helix-coiled-coil-helix domain containing 3 |
| chr12_+_112563303 | 0.18 |
ENST00000412615.2 |
TRAFD1 |
TRAF-type zinc finger domain containing 1 |
| chr13_-_41635512 | 0.18 |
ENST00000405737.2 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
| chr8_-_117886955 | 0.18 |
ENST00000297338.2 |
RAD21 |
RAD21 homolog (S. pombe) |
| chr7_+_87563557 | 0.18 |
ENST00000439864.1 ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22 |
ADAM metallopeptidase domain 22 |
| chr18_-_55470320 | 0.18 |
ENST00000536015.1 |
ATP8B1 |
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr5_-_102455801 | 0.17 |
ENST00000508629.1 ENST00000399004.2 |
GIN1 |
gypsy retrotransposon integrase 1 |
| chr15_+_57998923 | 0.17 |
ENST00000380557.4 |
POLR2M |
polymerase (RNA) II (DNA directed) polypeptide M |
| chrX_-_107334790 | 0.17 |
ENST00000217958.3 |
PSMD10 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chrX_+_9431324 | 0.17 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chrX_+_107334895 | 0.17 |
ENST00000372232.3 ENST00000345734.3 ENST00000372254.3 |
ATG4A |
autophagy related 4A, cysteine peptidase |
| chr2_-_90538397 | 0.17 |
ENST00000443397.3 |
RP11-685N3.1 |
Uncharacterized protein |
| chr3_+_135969148 | 0.17 |
ENST00000251654.4 ENST00000490504.1 ENST00000483687.1 ENST00000468777.1 ENST00000462637.1 ENST00000466072.1 ENST00000482086.1 ENST00000471595.1 ENST00000469217.1 ENST00000465423.1 ENST00000478469.1 |
PCCB |
propionyl CoA carboxylase, beta polypeptide |
| chr7_+_99102267 | 0.16 |
ENST00000326775.5 ENST00000451158.1 |
ZKSCAN5 |
zinc finger with KRAB and SCAN domains 5 |
| chr1_-_150669500 | 0.16 |
ENST00000271732.3 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr15_+_89787180 | 0.16 |
ENST00000300027.8 ENST00000310775.7 ENST00000567891.1 ENST00000564920.1 ENST00000565255.1 ENST00000567996.1 ENST00000451393.2 ENST00000563250.1 |
FANCI |
Fanconi anemia, complementation group I |
| chr16_-_31076273 | 0.16 |
ENST00000426488.2 |
ZNF668 |
zinc finger protein 668 |
| chr2_+_97203082 | 0.16 |
ENST00000454558.2 |
ARID5A |
AT rich interactive domain 5A (MRF1-like) |
| chr6_-_31648150 | 0.16 |
ENST00000375858.3 ENST00000383237.4 |
LY6G5C |
lymphocyte antigen 6 complex, locus G5C |
| chr2_-_70418032 | 0.16 |
ENST00000425268.1 ENST00000428751.1 ENST00000417203.1 ENST00000417865.1 ENST00000428010.1 ENST00000447804.1 ENST00000264434.2 |
C2orf42 |
chromosome 2 open reading frame 42 |
| chr8_-_41522779 | 0.16 |
ENST00000522231.1 ENST00000314214.8 ENST00000348036.4 ENST00000457297.1 ENST00000522543.1 |
ANK1 |
ankyrin 1, erythrocytic |
| chr1_+_155107820 | 0.16 |
ENST00000484157.1 |
SLC50A1 |
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr17_+_34848049 | 0.16 |
ENST00000588902.1 ENST00000591067.1 |
ZNHIT3 |
zinc finger, HIT-type containing 3 |
| chr3_-_105587879 | 0.16 |
ENST00000264122.4 ENST00000403724.1 ENST00000405772.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr10_+_60094735 | 0.16 |
ENST00000373910.4 |
UBE2D1 |
ubiquitin-conjugating enzyme E2D 1 |
| chr1_-_54872059 | 0.15 |
ENST00000371320.3 |
SSBP3 |
single stranded DNA binding protein 3 |
| chr8_-_71519889 | 0.15 |
ENST00000521425.1 |
TRAM1 |
translocation associated membrane protein 1 |
| chr11_-_64512469 | 0.15 |
ENST00000377485.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr1_+_172422026 | 0.15 |
ENST00000367725.4 |
C1orf105 |
chromosome 1 open reading frame 105 |
| chr1_+_12977513 | 0.15 |
ENST00000330881.5 |
PRAMEF7 |
PRAME family member 7 |
| chr11_-_82746587 | 0.15 |
ENST00000528379.1 ENST00000534103.1 |
RAB30 |
RAB30, member RAS oncogene family |
| chr19_+_16187085 | 0.15 |
ENST00000300933.4 |
TPM4 |
tropomyosin 4 |
| chr8_-_90769422 | 0.14 |
ENST00000524190.1 ENST00000523859.1 |
RP11-37B2.1 |
RP11-37B2.1 |
| chr19_-_1650666 | 0.14 |
ENST00000588136.1 |
TCF3 |
transcription factor 3 |
| chr8_-_41522719 | 0.14 |
ENST00000335651.6 |
ANK1 |
ankyrin 1, erythrocytic |
| chr8_-_82633476 | 0.14 |
ENST00000518419.1 ENST00000517588.1 ENST00000521895.1 ENST00000520076.1 ENST00000519523.1 ENST00000522520.1 ENST00000521287.1 ENST00000523096.1 ENST00000220669.5 ENST00000517450.1 |
ZFAND1 |
zinc finger, AN1-type domain 1 |
| chr3_+_52245458 | 0.14 |
ENST00000459884.1 |
ALAS1 |
aminolevulinate, delta-, synthase 1 |
| chr1_-_206785789 | 0.14 |
ENST00000437518.1 ENST00000367114.3 |
EIF2D |
eukaryotic translation initiation factor 2D |
| chr1_+_51701924 | 0.14 |
ENST00000242719.3 |
RNF11 |
ring finger protein 11 |
| chr1_+_202976493 | 0.14 |
ENST00000367242.3 |
TMEM183A |
transmembrane protein 183A |
| chr4_+_47487285 | 0.14 |
ENST00000273859.3 ENST00000504445.1 |
ATP10D |
ATPase, class V, type 10D |
| chrX_+_69509927 | 0.13 |
ENST00000374403.3 |
KIF4A |
kinesin family member 4A |
| chr1_-_155214436 | 0.13 |
ENST00000327247.5 |
GBA |
glucosidase, beta, acid |
| chr9_+_27109133 | 0.13 |
ENST00000519097.1 ENST00000380036.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chr6_-_27835357 | 0.13 |
ENST00000331442.3 |
HIST1H1B |
histone cluster 1, H1b |
| chr1_-_206785898 | 0.13 |
ENST00000271764.2 |
EIF2D |
eukaryotic translation initiation factor 2D |
| chr17_+_6347761 | 0.13 |
ENST00000250056.8 ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A |
family with sequence similarity 64, member A |
| chr17_-_7145106 | 0.13 |
ENST00000577035.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr6_+_31554779 | 0.13 |
ENST00000376090.2 |
LST1 |
leukocyte specific transcript 1 |
| chr19_+_16186903 | 0.12 |
ENST00000588507.1 |
TPM4 |
tropomyosin 4 |
| chr19_+_36486078 | 0.12 |
ENST00000378887.2 |
SDHAF1 |
succinate dehydrogenase complex assembly factor 1 |
| chr19_-_4535233 | 0.12 |
ENST00000381848.3 ENST00000588887.1 ENST00000586133.1 |
PLIN5 |
perilipin 5 |
| chr11_-_67169253 | 0.12 |
ENST00000527663.1 ENST00000312989.7 |
PPP1CA |
protein phosphatase 1, catalytic subunit, alpha isozyme |
| chr18_+_39535152 | 0.12 |
ENST00000262039.4 ENST00000398870.3 ENST00000586545.1 |
PIK3C3 |
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr1_-_149889382 | 0.12 |
ENST00000369145.1 ENST00000369146.3 |
SV2A |
synaptic vesicle glycoprotein 2A |
| chr15_-_83240507 | 0.12 |
ENST00000564522.1 ENST00000398592.2 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr16_-_31105870 | 0.12 |
ENST00000394971.3 |
VKORC1 |
vitamin K epoxide reductase complex, subunit 1 |
| chr20_-_23030296 | 0.11 |
ENST00000377103.2 |
THBD |
thrombomodulin |
| chr18_+_158513 | 0.11 |
ENST00000400266.3 ENST00000580410.1 ENST00000383589.2 ENST00000261601.7 |
USP14 |
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) |
| chr1_+_171154347 | 0.11 |
ENST00000209929.7 ENST00000441535.1 |
FMO2 |
flavin containing monooxygenase 2 (non-functional) |
| chrX_+_103810874 | 0.11 |
ENST00000372582.1 |
IL1RAPL2 |
interleukin 1 receptor accessory protein-like 2 |
| chr19_+_18699599 | 0.11 |
ENST00000450195.2 |
C19orf60 |
chromosome 19 open reading frame 60 |
| chr14_-_50154921 | 0.11 |
ENST00000553805.2 ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2 |
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr16_-_31106048 | 0.11 |
ENST00000300851.6 |
VKORC1 |
vitamin K epoxide reductase complex, subunit 1 |
| chr6_-_13711773 | 0.11 |
ENST00000011619.3 |
RANBP9 |
RAN binding protein 9 |
| chr9_-_123239632 | 0.11 |
ENST00000416449.1 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr2_+_54198210 | 0.11 |
ENST00000607452.1 ENST00000422521.2 |
ACYP2 |
acylphosphatase 2, muscle type |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.4 | 1.7 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.2 | 0.9 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.2 | 0.8 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.2 | 1.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.9 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 1.3 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.4 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 1.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 0.7 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.3 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 2.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.3 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 0.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.8 | GO:0008061 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.1 | 0.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 1.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 2.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.3 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.5 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 1.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.0 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 1.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.0 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.0 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.0 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 2.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.9 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.9 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | GO:0097319 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.4 | 1.1 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.3 | 1.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.2 | 1.1 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.2 | 0.8 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.2 | 0.7 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.2 | 1.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.9 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.4 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 1.0 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.1 | 0.4 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.9 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 1.0 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.3 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.4 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.8 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.2 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 0.6 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 1.3 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 0.4 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 | 0.8 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.2 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 0.2 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 1.8 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.1 | 0.2 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 0.5 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.1 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.0 | 0.5 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.3 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:1904457 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0070781 | response to biotin(GO:0070781) |
| 0.0 | 0.3 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.1 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.8 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 2.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.2 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.0 | 0.3 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.1 | GO:0002884 | regulation of type IV hypersensitivity(GO:0001807) negative regulation of hypersensitivity(GO:0002884) |
| 0.0 | 0.1 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) malonyl-CoA metabolic process(GO:2001293) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.3 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 1.8 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 2.0 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 1.0 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 1.0 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.0 | GO:0043012 | regulation of fusion of sperm to egg plasma membrane(GO:0043012) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.7 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) |
| 0.0 | 0.1 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.0 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0002169 | 3-methylcrotonyl-CoA carboxylase complex, mitochondrial(GO:0002169) methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 0.4 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 1.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.7 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.4 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.3 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 2.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 1.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.6 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 1.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 3.1 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 1.1 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.1 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |