Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
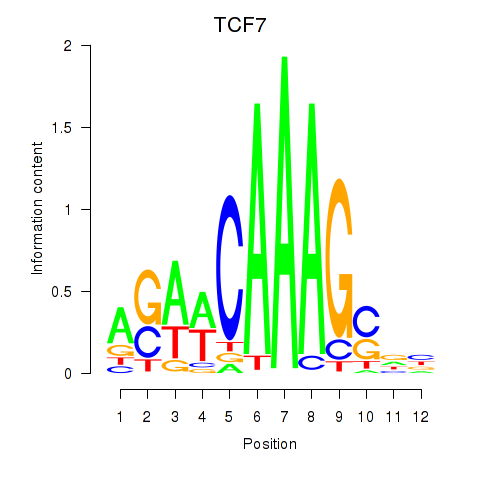
Results for TCF7
Z-value: 1.15
Transcription factors associated with TCF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7
|
ENSG00000081059.15 | TCF7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7 | hg19_v2_chr5_+_133477905_133477956 | -0.76 | 5.8e-04 | Click! |
Activity profile of TCF7 motif
Sorted Z-values of TCF7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_151344172 | 7.50 |
ENST00000375734.2 ENST00000263895.4 ENST00000454202.1 |
RND3 |
Rho family GTPase 3 |
| chr10_+_54074033 | 4.34 |
ENST00000373970.3 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
| chr7_+_94023873 | 3.74 |
ENST00000297268.6 |
COL1A2 |
collagen, type I, alpha 2 |
| chr12_+_6309963 | 3.67 |
ENST00000382515.2 |
CD9 |
CD9 molecule |
| chr5_+_34757309 | 3.60 |
ENST00000397449.1 |
RAI14 |
retinoic acid induced 14 |
| chr8_-_119964434 | 3.56 |
ENST00000297350.4 |
TNFRSF11B |
tumor necrosis factor receptor superfamily, member 11b |
| chr5_-_16936340 | 3.07 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr3_+_159570722 | 2.65 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr15_+_74218787 | 2.53 |
ENST00000261921.7 |
LOXL1 |
lysyl oxidase-like 1 |
| chr7_-_80548667 | 2.44 |
ENST00000265361.3 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr5_+_135364584 | 2.26 |
ENST00000442011.2 ENST00000305126.8 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr7_-_107643674 | 2.17 |
ENST00000222399.6 |
LAMB1 |
laminin, beta 1 |
| chr1_-_79472365 | 2.15 |
ENST00000370742.3 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr14_+_85996471 | 2.15 |
ENST00000330753.4 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr9_-_35691017 | 2.04 |
ENST00000378292.3 |
TPM2 |
tropomyosin 2 (beta) |
| chr7_+_134576317 | 2.01 |
ENST00000424922.1 ENST00000495522.1 |
CALD1 |
caldesmon 1 |
| chr9_-_130639997 | 1.96 |
ENST00000373176.1 |
AK1 |
adenylate kinase 1 |
| chr5_-_157002775 | 1.87 |
ENST00000257527.4 |
ADAM19 |
ADAM metallopeptidase domain 19 |
| chr10_+_17271266 | 1.79 |
ENST00000224237.5 |
VIM |
vimentin |
| chr2_+_234621551 | 1.78 |
ENST00000608381.1 ENST00000373414.3 |
UGT1A1 UGT1A5 |
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr7_+_130131907 | 1.78 |
ENST00000223215.4 ENST00000437945.1 |
MEST |
mesoderm specific transcript |
| chr8_+_30244580 | 1.74 |
ENST00000523115.1 ENST00000519647.1 |
RBPMS |
RNA binding protein with multiple splicing |
| chr7_+_134576151 | 1.72 |
ENST00000393118.2 |
CALD1 |
caldesmon 1 |
| chr13_+_102142296 | 1.64 |
ENST00000376162.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr2_-_217560248 | 1.61 |
ENST00000233813.4 |
IGFBP5 |
insulin-like growth factor binding protein 5 |
| chr1_-_110933663 | 1.58 |
ENST00000369781.4 ENST00000541986.1 ENST00000369779.4 |
SLC16A4 |
solute carrier family 16, member 4 |
| chr1_-_110933611 | 1.50 |
ENST00000472422.2 ENST00000437429.2 |
SLC16A4 |
solute carrier family 16, member 4 |
| chr13_+_102104980 | 1.45 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr6_+_83073334 | 1.43 |
ENST00000369750.3 |
TPBG |
trophoblast glycoprotein |
| chr4_-_52904425 | 1.43 |
ENST00000535450.1 |
SGCB |
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr17_-_46623441 | 1.39 |
ENST00000330070.4 |
HOXB2 |
homeobox B2 |
| chr13_+_102104952 | 1.37 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr6_-_112575912 | 1.37 |
ENST00000522006.1 ENST00000230538.7 ENST00000519932.1 |
LAMA4 |
laminin, alpha 4 |
| chr3_+_138067521 | 1.33 |
ENST00000494949.1 |
MRAS |
muscle RAS oncogene homolog |
| chrX_+_51927919 | 1.25 |
ENST00000416960.1 |
MAGED4 |
melanoma antigen family D, 4 |
| chr12_-_10022735 | 1.25 |
ENST00000228438.2 |
CLEC2B |
C-type lectin domain family 2, member B |
| chr6_+_83073952 | 1.21 |
ENST00000543496.1 |
TPBG |
trophoblast glycoprotein |
| chr6_-_112575687 | 1.20 |
ENST00000521398.1 ENST00000424408.2 ENST00000243219.3 |
LAMA4 |
laminin, alpha 4 |
| chr8_-_145047688 | 1.19 |
ENST00000356346.3 |
PLEC |
plectin |
| chr7_-_27213893 | 1.18 |
ENST00000283921.4 |
HOXA10 |
homeobox A10 |
| chr22_+_39052632 | 1.14 |
ENST00000411557.1 ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1 |
chibby homolog 1 (Drosophila) |
| chr14_+_85996507 | 1.13 |
ENST00000554746.1 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr9_+_124088860 | 1.12 |
ENST00000373806.1 |
GSN |
gelsolin |
| chr4_-_7873981 | 1.05 |
ENST00000360265.4 |
AFAP1 |
actin filament associated protein 1 |
| chr12_+_21654714 | 1.01 |
ENST00000542038.1 ENST00000540141.1 ENST00000229314.5 |
GOLT1B |
golgi transport 1B |
| chr7_+_23286182 | 0.99 |
ENST00000258733.4 ENST00000381990.2 ENST00000409458.3 ENST00000539136.1 ENST00000453162.2 |
GPNMB |
glycoprotein (transmembrane) nmb |
| chr2_+_46926326 | 0.98 |
ENST00000394861.2 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr2_+_189157536 | 0.93 |
ENST00000409580.1 ENST00000409637.3 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
| chr2_-_36825281 | 0.92 |
ENST00000405912.3 ENST00000379245.4 |
FEZ2 |
fasciculation and elongation protein zeta 2 (zygin II) |
| chr4_+_87515454 | 0.91 |
ENST00000427191.2 ENST00000436978.1 ENST00000502971.1 |
PTPN13 |
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) |
| chr1_-_209824643 | 0.91 |
ENST00000391911.1 ENST00000415782.1 |
LAMB3 |
laminin, beta 3 |
| chr2_-_231989808 | 0.90 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr4_+_154125565 | 0.88 |
ENST00000338700.5 |
TRIM2 |
tripartite motif containing 2 |
| chr17_-_7297519 | 0.84 |
ENST00000576362.1 ENST00000571078.1 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr11_-_33774944 | 0.84 |
ENST00000532057.1 ENST00000531080.1 |
FBXO3 |
F-box protein 3 |
| chr2_+_69240302 | 0.83 |
ENST00000303714.4 |
ANTXR1 |
anthrax toxin receptor 1 |
| chr6_-_112575838 | 0.83 |
ENST00000455073.1 |
LAMA4 |
laminin, alpha 4 |
| chr18_+_3449821 | 0.80 |
ENST00000407501.2 ENST00000405385.3 ENST00000546979.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr8_-_101962777 | 0.79 |
ENST00000395951.3 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chrX_+_54835493 | 0.78 |
ENST00000396224.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr14_+_74486043 | 0.76 |
ENST00000464394.1 ENST00000394009.3 |
CCDC176 |
coiled-coil domain containing 176 |
| chrX_+_12993336 | 0.76 |
ENST00000380635.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr9_+_125137565 | 0.74 |
ENST00000373698.5 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr9_-_132586378 | 0.73 |
ENST00000351698.4 |
TOR1A |
torsin family 1, member A (torsin A) |
| chr6_+_52285046 | 0.72 |
ENST00000371068.5 |
EFHC1 |
EF-hand domain (C-terminal) containing 1 |
| chr1_-_151345159 | 0.70 |
ENST00000458566.1 ENST00000447402.3 ENST00000426705.2 ENST00000435071.1 ENST00000368868.5 |
SELENBP1 |
selenium binding protein 1 |
| chr4_-_129209944 | 0.69 |
ENST00000520121.1 |
PGRMC2 |
progesterone receptor membrane component 2 |
| chr11_+_65339820 | 0.68 |
ENST00000316409.2 ENST00000449319.2 ENST00000530349.1 |
FAM89B |
family with sequence similarity 89, member B |
| chr11_+_7506713 | 0.67 |
ENST00000329293.3 ENST00000534244.1 |
OLFML1 |
olfactomedin-like 1 |
| chr13_-_36920872 | 0.67 |
ENST00000451493.1 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr6_-_112575758 | 0.65 |
ENST00000431543.2 ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4 |
laminin, alpha 4 |
| chr13_-_36920615 | 0.65 |
ENST00000494062.2 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr19_+_1026566 | 0.62 |
ENST00000348419.3 ENST00000565096.2 ENST00000562958.2 ENST00000562075.2 ENST00000607102.1 |
CNN2 |
calponin 2 |
| chr16_+_68678739 | 0.61 |
ENST00000264012.4 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr8_-_98290087 | 0.59 |
ENST00000322128.3 |
TSPYL5 |
TSPY-like 5 |
| chr2_-_208031943 | 0.59 |
ENST00000421199.1 ENST00000457962.1 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr13_-_36920420 | 0.58 |
ENST00000438666.2 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr15_+_74466012 | 0.57 |
ENST00000249842.3 |
ISLR |
immunoglobulin superfamily containing leucine-rich repeat |
| chr5_-_157002749 | 0.57 |
ENST00000517905.1 ENST00000430702.2 ENST00000394020.1 |
ADAM19 |
ADAM metallopeptidase domain 19 |
| chrX_-_13956737 | 0.55 |
ENST00000454189.2 |
GPM6B |
glycoprotein M6B |
| chrX_+_102883620 | 0.52 |
ENST00000372626.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chrX_+_12993202 | 0.51 |
ENST00000451311.2 ENST00000380636.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr11_-_65625678 | 0.51 |
ENST00000308162.5 |
CFL1 |
cofilin 1 (non-muscle) |
| chr5_-_140700322 | 0.50 |
ENST00000313368.5 |
TAF7 |
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
| chr2_+_181845298 | 0.49 |
ENST00000410062.4 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
| chr21_+_39628852 | 0.46 |
ENST00000398938.2 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr1_+_68150744 | 0.46 |
ENST00000370986.4 ENST00000370985.3 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
| chr10_-_49860525 | 0.46 |
ENST00000435790.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr11_-_64013663 | 0.46 |
ENST00000392210.2 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr17_-_79818354 | 0.45 |
ENST00000576541.1 ENST00000576380.1 ENST00000571617.1 ENST00000576052.1 ENST00000576390.1 ENST00000573778.2 ENST00000439918.2 ENST00000574914.1 ENST00000331483.4 |
P4HB |
prolyl 4-hydroxylase, beta polypeptide |
| chr1_-_161993616 | 0.45 |
ENST00000294794.3 |
OLFML2B |
olfactomedin-like 2B |
| chr19_+_1026298 | 0.43 |
ENST00000263097.4 |
CNN2 |
calponin 2 |
| chr9_+_91003271 | 0.42 |
ENST00000375859.3 ENST00000541629.1 |
SPIN1 |
spindlin 1 |
| chr12_+_4714145 | 0.42 |
ENST00000545342.1 |
DYRK4 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr19_+_45281118 | 0.41 |
ENST00000270279.3 ENST00000341505.4 |
CBLC |
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr8_-_17579726 | 0.41 |
ENST00000381861.3 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr3_+_35681081 | 0.40 |
ENST00000428373.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr13_+_29233218 | 0.39 |
ENST00000380842.4 |
POMP |
proteasome maturation protein |
| chr9_+_125703282 | 0.38 |
ENST00000373647.4 ENST00000402311.1 |
RABGAP1 |
RAB GTPase activating protein 1 |
| chr17_-_26989136 | 0.38 |
ENST00000247020.4 |
SDF2 |
stromal cell-derived factor 2 |
| chrX_+_135618258 | 0.37 |
ENST00000440515.1 ENST00000456412.1 |
VGLL1 |
vestigial like 1 (Drosophila) |
| chr1_-_16345245 | 0.37 |
ENST00000311890.9 |
HSPB7 |
heat shock 27kDa protein family, member 7 (cardiovascular) |
| chrX_-_106960285 | 0.37 |
ENST00000503515.1 ENST00000372397.2 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr5_-_148930960 | 0.37 |
ENST00000261798.5 ENST00000377843.2 |
CSNK1A1 |
casein kinase 1, alpha 1 |
| chr2_+_10183651 | 0.36 |
ENST00000305883.1 |
KLF11 |
Kruppel-like factor 11 |
| chr12_-_133405288 | 0.36 |
ENST00000204726.3 |
GOLGA3 |
golgin A3 |
| chr3_+_36421826 | 0.36 |
ENST00000273183.3 |
STAC |
SH3 and cysteine rich domain |
| chrX_-_148676974 | 0.35 |
ENST00000524178.1 |
HSFX2 |
heat shock transcription factor family, X linked 2 |
| chr10_+_52750930 | 0.34 |
ENST00000401604.2 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr22_+_19950060 | 0.34 |
ENST00000449653.1 |
COMT |
catechol-O-methyltransferase |
| chr1_-_153113927 | 0.34 |
ENST00000368752.4 |
SPRR2B |
small proline-rich protein 2B |
| chr21_+_39628655 | 0.33 |
ENST00000398925.1 ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr3_+_111717600 | 0.33 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr3_-_125775629 | 0.33 |
ENST00000383598.2 |
SLC41A3 |
solute carrier family 41, member 3 |
| chr2_-_86564776 | 0.33 |
ENST00000165698.5 ENST00000541910.1 ENST00000535845.1 |
REEP1 |
receptor accessory protein 1 |
| chr8_+_73921085 | 0.32 |
ENST00000276603.5 ENST00000276602.6 ENST00000518874.1 |
TERF1 |
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr13_+_45694583 | 0.31 |
ENST00000340473.6 |
GTF2F2 |
general transcription factor IIF, polypeptide 2, 30kDa |
| chr5_-_82969405 | 0.31 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chr9_-_100459639 | 0.31 |
ENST00000375128.4 |
XPA |
xeroderma pigmentosum, complementation group A |
| chr17_+_48610074 | 0.31 |
ENST00000503690.1 ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3 |
epsin 3 |
| chr6_+_125540951 | 0.31 |
ENST00000524679.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr3_+_111717511 | 0.31 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr12_-_6055398 | 0.31 |
ENST00000327087.8 ENST00000356134.5 ENST00000546188.1 |
ANO2 |
anoctamin 2 |
| chrX_+_148855726 | 0.31 |
ENST00000370416.4 |
HSFX1 |
heat shock transcription factor family, X linked 1 |
| chr19_+_13135386 | 0.31 |
ENST00000360105.4 ENST00000588228.1 ENST00000591028.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr12_-_10151773 | 0.30 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr1_-_153013588 | 0.30 |
ENST00000360379.3 |
SPRR2D |
small proline-rich protein 2D |
| chr20_+_31823792 | 0.30 |
ENST00000375413.4 ENST00000354297.4 ENST00000375422.2 |
BPIFA1 |
BPI fold containing family A, member 1 |
| chr4_+_159593271 | 0.30 |
ENST00000512251.1 ENST00000511912.1 |
ETFDH |
electron-transferring-flavoprotein dehydrogenase |
| chr14_+_61201445 | 0.30 |
ENST00000261245.4 ENST00000539616.2 |
MNAT1 |
MNAT CDK-activating kinase assembly factor 1 |
| chr10_+_52833883 | 0.29 |
ENST00000373980.4 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr19_+_58898627 | 0.29 |
ENST00000598098.1 ENST00000598495.1 ENST00000196551.3 ENST00000596046.1 |
RPS5 |
ribosomal protein S5 |
| chrX_+_100663243 | 0.28 |
ENST00000316594.5 |
HNRNPH2 |
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr5_+_140165876 | 0.28 |
ENST00000504120.2 ENST00000394633.3 ENST00000378133.3 |
PCDHA1 |
protocadherin alpha 1 |
| chr7_+_80231466 | 0.27 |
ENST00000309881.7 ENST00000534394.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr9_-_21187598 | 0.27 |
ENST00000421715.1 |
IFNA4 |
interferon, alpha 4 |
| chr2_+_234580525 | 0.27 |
ENST00000609637.1 |
UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr6_-_32145861 | 0.26 |
ENST00000336984.6 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr11_-_122930121 | 0.25 |
ENST00000524552.1 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr19_+_42829702 | 0.25 |
ENST00000334370.4 |
MEGF8 |
multiple EGF-like-domains 8 |
| chr17_-_37009882 | 0.24 |
ENST00000378096.3 ENST00000394332.1 ENST00000394333.1 ENST00000577407.1 ENST00000479035.2 |
RPL23 |
ribosomal protein L23 |
| chr19_+_41256764 | 0.24 |
ENST00000243563.3 ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA |
small nuclear ribonucleoprotein polypeptide A |
| chr2_+_234580499 | 0.23 |
ENST00000354728.4 |
UGT1A9 |
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chrX_-_55020511 | 0.23 |
ENST00000375006.3 ENST00000374992.2 |
PFKFB1 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr8_-_20161466 | 0.23 |
ENST00000381569.1 |
LZTS1 |
leucine zipper, putative tumor suppressor 1 |
| chr3_-_126327398 | 0.22 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chrX_-_153599578 | 0.22 |
ENST00000360319.4 ENST00000344736.4 |
FLNA |
filamin A, alpha |
| chr3_+_118892362 | 0.22 |
ENST00000497685.1 ENST00000264234.3 |
UPK1B |
uroplakin 1B |
| chr2_+_46926048 | 0.21 |
ENST00000306503.5 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr1_-_153029980 | 0.21 |
ENST00000392653.2 |
SPRR2A |
small proline-rich protein 2A |
| chr4_-_74486347 | 0.20 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr11_-_64013288 | 0.20 |
ENST00000542235.1 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr2_+_27593389 | 0.20 |
ENST00000233575.2 ENST00000543024.1 ENST00000537606.1 |
SNX17 |
sorting nexin 17 |
| chr22_+_39077947 | 0.20 |
ENST00000216034.4 |
TOMM22 |
translocase of outer mitochondrial membrane 22 homolog (yeast) |
| chr14_+_101299520 | 0.19 |
ENST00000455531.1 |
MEG3 |
maternally expressed 3 (non-protein coding) |
| chr11_-_6677018 | 0.18 |
ENST00000299441.3 |
DCHS1 |
dachsous cadherin-related 1 |
| chr7_+_55086703 | 0.18 |
ENST00000455089.1 ENST00000342916.3 ENST00000344576.2 ENST00000420316.2 |
EGFR |
epidermal growth factor receptor |
| chr4_-_139163491 | 0.18 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr5_+_140579162 | 0.17 |
ENST00000536699.1 ENST00000354757.3 |
PCDHB11 |
protocadherin beta 11 |
| chr17_-_17485731 | 0.17 |
ENST00000395783.1 |
PEMT |
phosphatidylethanolamine N-methyltransferase |
| chr10_+_99332198 | 0.17 |
ENST00000307518.5 ENST00000298808.5 ENST00000370655.1 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr1_-_211752073 | 0.17 |
ENST00000367001.4 |
SLC30A1 |
solute carrier family 30 (zinc transporter), member 1 |
| chr3_-_167813672 | 0.16 |
ENST00000470487.1 |
GOLIM4 |
golgi integral membrane protein 4 |
| chr11_-_10829851 | 0.16 |
ENST00000532082.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr19_-_54746600 | 0.16 |
ENST00000245621.5 ENST00000270464.5 ENST00000419410.2 ENST00000391735.3 ENST00000396365.2 ENST00000440558.2 ENST00000407860.2 |
LILRA6 LILRB3 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6 leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 |
| chr22_-_42526802 | 0.15 |
ENST00000359033.4 ENST00000389970.3 ENST00000360608.5 |
CYP2D6 |
cytochrome P450, family 2, subfamily D, polypeptide 6 |
| chr1_-_149900122 | 0.15 |
ENST00000271628.8 |
SF3B4 |
splicing factor 3b, subunit 4, 49kDa |
| chr2_-_136288113 | 0.14 |
ENST00000401392.1 |
ZRANB3 |
zinc finger, RAN-binding domain containing 3 |
| chr5_-_137610300 | 0.14 |
ENST00000274721.3 |
GFRA3 |
GDNF family receptor alpha 3 |
| chr9_-_127533519 | 0.14 |
ENST00000487099.2 ENST00000344523.4 ENST00000373584.3 |
NR6A1 |
nuclear receptor subfamily 6, group A, member 1 |
| chr3_+_111718036 | 0.13 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr12_-_89920030 | 0.13 |
ENST00000413530.1 ENST00000547474.1 |
GALNT4 POC1B-GALNT4 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr7_-_142207004 | 0.13 |
ENST00000426318.2 |
TRBV10-2 |
T cell receptor beta variable 10-2 |
| chr2_-_27593306 | 0.12 |
ENST00000347454.4 |
EIF2B4 |
eukaryotic translation initiation factor 2B, subunit 4 delta, 67kDa |
| chr11_+_113848216 | 0.12 |
ENST00000299961.5 |
HTR3A |
5-hydroxytryptamine (serotonin) receptor 3A, ionotropic |
| chr19_+_57791419 | 0.12 |
ENST00000537645.1 |
ZNF460 |
zinc finger protein 460 |
| chr15_+_52043758 | 0.12 |
ENST00000249700.4 ENST00000539962.2 |
TMOD2 |
tropomodulin 2 (neuronal) |
| chr1_+_59762642 | 0.11 |
ENST00000371218.4 ENST00000303721.7 |
FGGY |
FGGY carbohydrate kinase domain containing |
| chr11_-_10830463 | 0.11 |
ENST00000527419.1 ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr8_+_133879193 | 0.11 |
ENST00000377869.1 ENST00000220616.4 |
TG |
thyroglobulin |
| chr8_-_6914251 | 0.11 |
ENST00000330590.2 |
DEFA5 |
defensin, alpha 5, Paneth cell-specific |
| chr22_-_37880543 | 0.10 |
ENST00000442496.1 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr5_-_137610254 | 0.10 |
ENST00000378362.3 |
GFRA3 |
GDNF family receptor alpha 3 |
| chr4_-_159080806 | 0.09 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr17_-_56065484 | 0.09 |
ENST00000581208.1 |
VEZF1 |
vascular endothelial zinc finger 1 |
| chr4_-_5891918 | 0.09 |
ENST00000512574.1 |
CRMP1 |
collapsin response mediator protein 1 |
| chr19_-_40791211 | 0.08 |
ENST00000579047.1 |
AKT2 |
v-akt murine thymoma viral oncogene homolog 2 |
| chr12_+_27677085 | 0.08 |
ENST00000545334.1 ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr3_-_197676740 | 0.08 |
ENST00000452735.1 ENST00000453254.1 ENST00000455191.1 |
IQCG |
IQ motif containing G |
| chr5_-_157286104 | 0.08 |
ENST00000530742.1 ENST00000523908.1 ENST00000523094.1 ENST00000296951.5 ENST00000411809.2 |
CLINT1 |
clathrin interactor 1 |
| chr14_+_21214039 | 0.08 |
ENST00000326842.2 |
EDDM3A |
epididymal protein 3A |
| chr1_+_10459111 | 0.08 |
ENST00000541529.1 ENST00000270776.8 ENST00000483936.1 ENST00000538557.1 |
PGD |
phosphogluconate dehydrogenase |
| chr2_+_1417228 | 0.07 |
ENST00000382269.3 ENST00000337415.3 ENST00000345913.4 ENST00000346956.3 ENST00000349624.3 ENST00000539820.1 ENST00000329066.4 ENST00000382201.3 |
TPO |
thyroid peroxidase |
| chr11_-_129062093 | 0.07 |
ENST00000310343.9 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr9_-_125675576 | 0.07 |
ENST00000373659.3 |
ZBTB6 |
zinc finger and BTB domain containing 6 |
| chr20_-_17511962 | 0.06 |
ENST00000377873.3 |
BFSP1 |
beaded filament structural protein 1, filensin |
| chr8_+_145064215 | 0.06 |
ENST00000313269.5 |
GRINA |
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr2_+_7017796 | 0.06 |
ENST00000382040.3 |
RSAD2 |
radical S-adenosyl methionine domain containing 2 |
| chrX_-_102757802 | 0.06 |
ENST00000372633.1 |
RAB40A |
RAB40A, member RAS oncogene family |
| chr2_-_166060571 | 0.06 |
ENST00000360093.3 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.3 | 0.9 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 1.7 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 1.0 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.2 | 0.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 0.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 0.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.7 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 0.9 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.1 | 0.8 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.9 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 1.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.2 | GO:0044094 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.1 | 0.6 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.7 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.8 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 7.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 1.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.5 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.2 | GO:1990909 | catenin complex(GO:0016342) Wnt signalosome(GO:1990909) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 2.9 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 5.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 10.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.2 | 4.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 4.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 5.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 11.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 5.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 1.6 | 4.8 | GO:0009996 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) negative regulation of cell fate specification(GO:0009996) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 1.2 | 4.7 | GO:0018032 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 1.1 | 6.6 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.7 | 6.5 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.6 | 2.3 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.6 | 2.8 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.5 | 1.5 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.5 | 6.0 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.4 | 5.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.4 | 0.8 | GO:1904833 | positive regulation of superoxide dismutase activity(GO:1901671) positive regulation of removal of superoxide radicals(GO:1904833) |
| 0.3 | 2.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.3 | 0.9 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.3 | 1.8 | GO:2000334 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.3 | 0.8 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) response to antineoplastic agent(GO:0097327) |
| 0.2 | 0.7 | GO:0061055 | myotome development(GO:0061055) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 0.7 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.2 | 5.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.2 | 10.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.2 | 0.7 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 10.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 1.0 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.7 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.1 | 0.5 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 1.2 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 1.0 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 1.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.3 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 2.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 1.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 1.0 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 3.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.6 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.2 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.2 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 0.6 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.0 | 5.8 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.8 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 10.0 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 3.0 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 3.8 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.3 | GO:0032196 | transposition(GO:0032196) |
| 0.0 | 1.3 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.6 | GO:0001885 | endothelial cell development(GO:0001885) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) negative regulation of cell fate specification(GO:0009996) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.9 | 3.6 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.5 | 3.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.5 | 2.2 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.5 | 1.6 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.5 | 1.9 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.5 | 1.4 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.4 | 2.0 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.4 | 2.4 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.3 | 3.3 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.3 | 0.8 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.2 | 3.7 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.2 | 1.1 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.2 | 1.3 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.2 | 0.6 | GO:0032771 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.2 | 1.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 0.9 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.2 | 0.7 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.2 | 1.8 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.2 | 0.9 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.7 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.1 | 0.8 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 1.4 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 0.3 | GO:0031627 | telomeric loop formation(GO:0031627) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.1 | 0.5 | GO:2000782 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) regulation of barbed-end actin filament capping(GO:2000812) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.1 | 1.7 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 0.3 | GO:1900229 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.1 | 2.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0031337 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.1 | 0.3 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) fasciculation of sensory neuron axon(GO:0097155) |
| 0.1 | 0.2 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.2 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.1 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.2 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 0.6 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.8 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.4 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.1 | 2.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.2 | GO:0033076 | isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.1 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.3 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.1 | 0.5 | GO:0015846 | polyamine transport(GO:0015846) |
| 0.0 | 0.2 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.2 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.4 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 2.7 | GO:0035904 | aorta development(GO:0035904) |
| 0.0 | 0.6 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 1.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0033133 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 2.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 2.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 4.1 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.2 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:1904616 | regulation of actin filament binding(GO:1904529) negative regulation of actin filament binding(GO:1904530) regulation of actin binding(GO:1904616) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.5 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.3 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 1.2 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.7 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 2.2 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 1.4 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.1 | GO:1903960 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) |
| 0.0 | 0.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.2 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.0 | 0.2 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.8 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 1.1 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 1.0 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 0.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.8 | GO:0055007 | cardiac muscle cell differentiation(GO:0055007) |
| 0.0 | 0.6 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.4 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.9 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 1.6 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0019321 | pentose metabolic process(GO:0019321) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.6 | 1.7 | GO:0051300 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.5 | 1.6 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.4 | 1.3 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.4 | 1.4 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.3 | 1.5 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.3 | 1.7 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.3 | 0.8 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.3 | 0.8 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.2 | 1.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.2 | 0.9 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 1.0 | GO:0044336 | embryonic genitalia morphogenesis(GO:0030538) canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.2 | 0.8 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.2 | 0.6 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.2 | 0.6 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.2 | 0.9 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.2 | 1.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 0.9 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.2 | 0.5 | GO:1902356 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.2 | 0.6 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.2 | 1.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 0.6 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 1.0 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.6 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.7 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 1.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 2.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.5 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 | 0.4 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.2 | GO:1905064 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 4.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.8 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.4 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.3 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.5 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.7 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.7 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.2 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.3 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.7 | GO:0000165 | MAPK cascade(GO:0000165) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.4 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.6 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.7 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.6 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0060339 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.3 | GO:0051005 | endosome transport via multivesicular body sorting pathway(GO:0032509) negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 1.4 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 1.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.0 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.5 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.5 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.3 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.7 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.4 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.8 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.2 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.3 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.2 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.7 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.4 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.3 | 4.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 3.7 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 1.6 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 1.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 2.7 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 3.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.7 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.4 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 2.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.9 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 2.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 3.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.3 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.2 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.1 | 1.9 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 4.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.3 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.3 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 1.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 4.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 16.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.3 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 3.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.0 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 1.2 | 4.7 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 1.1 | 10.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.5 | 1.5 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.4 | 4.8 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 1.8 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.3 | 5.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 1.2 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.2 | 4.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 6.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 3.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 0.9 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 1.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.6 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 3.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 6.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.8 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 0.3 | GO:0004803 | transposase activity(GO:0004803) |
| 0.1 | 7.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 5.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 5.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 1.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.6 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 4.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.5 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 8.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.7 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 1.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.2 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 3.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.3 | 3.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 1.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 1.9 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 3.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 2.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 2.5 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 1.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 3.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.6 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.5 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 1.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 1.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 2.9 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 1.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.9 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 1.0 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.1 | 6.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 2.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.2 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 3.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.7 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 1.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.4 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 1.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 1.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 2.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 1.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0017057 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 4.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 1.4 | GO:0005178 | integrin binding(GO:0005178) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.6 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.5 | 10.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.4 | 1.4 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.2 | 1.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 0.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 2.8 | GO:0043203 | axon hillock(GO:0043203) |
| 0.2 | 4.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 4.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 1.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 13.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 2.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 4.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 10.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 1.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 4.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 10.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 2.2 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 3.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 3.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 10.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.3 | 6.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 4.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 10.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.7 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 5.6 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 2.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 4.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 5.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 3.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.9 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 4.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 4.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 4.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.0 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 2.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 5.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 5.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 2.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.5 | 1.6 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.3 | 0.9 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.3 | 2.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 1.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 0.9 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.2 | 1.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.6 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.2 | 0.6 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.2 | 0.9 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.2 | 0.5 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.2 | 0.6 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 0.4 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 2.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 3.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.7 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 0.4 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 1.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 1.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.3 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.3 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 5.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.8 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.5 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) primary miRNA binding(GO:0070878) |
| 0.0 | 0.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 2.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.5 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |