Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
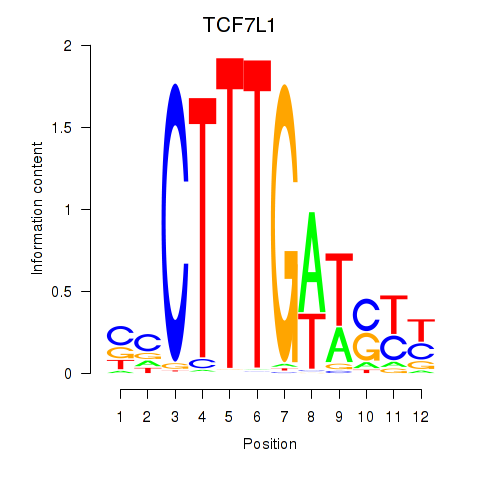
Results for TCF7L1
Z-value: 0.97
Transcription factors associated with TCF7L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7L1
|
ENSG00000152284.4 | TCF7L1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TCF7L1 | hg19_v2_chr2_+_85360499_85360598 | -0.70 | 2.4e-03 | Click! |
Activity profile of TCF7L1 motif
Sorted Z-values of TCF7L1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TCF7L1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_65409502 | 3.15 |
ENST00000389614.5 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr3_-_148939835 | 2.60 |
ENST00000264613.6 |
CP |
ceruloplasmin (ferroxidase) |
| chr10_+_7745232 | 2.13 |
ENST00000358415.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_+_7745303 | 2.12 |
ENST00000429820.1 ENST00000379587.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr5_-_175964366 | 2.04 |
ENST00000274811.4 |
RNF44 |
ring finger protein 44 |
| chr2_+_228678550 | 1.60 |
ENST00000409189.3 ENST00000358813.4 |
CCL20 |
chemokine (C-C motif) ligand 20 |
| chr6_-_29527702 | 1.55 |
ENST00000377050.4 |
UBD |
ubiquitin D |
| chr16_-_73082274 | 1.43 |
ENST00000268489.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr1_+_200993071 | 1.38 |
ENST00000446333.1 ENST00000458003.1 |
RP11-168O16.1 |
RP11-168O16.1 |
| chr14_-_23451467 | 1.29 |
ENST00000555074.1 ENST00000361265.4 |
RP11-298I3.5 AJUBA |
RP11-298I3.5 ajuba LIM protein |
| chr17_-_39093672 | 1.15 |
ENST00000209718.3 ENST00000436344.3 ENST00000485751.1 |
KRT23 |
keratin 23 (histone deacetylase inducible) |
| chr4_+_113152881 | 1.08 |
ENST00000274000.5 |
AP1AR |
adaptor-related protein complex 1 associated regulatory protein |
| chr5_+_133450365 | 1.04 |
ENST00000342854.5 ENST00000321603.6 ENST00000321584.4 ENST00000378564.1 ENST00000395029.1 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
| chr4_+_113152978 | 1.02 |
ENST00000309703.6 |
AP1AR |
adaptor-related protein complex 1 associated regulatory protein |
| chr2_+_11679963 | 1.02 |
ENST00000263834.5 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr15_+_43985084 | 1.00 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr11_-_87908600 | 0.99 |
ENST00000531138.1 ENST00000526372.1 ENST00000243662.6 |
RAB38 |
RAB38, member RAS oncogene family |
| chr15_+_43885252 | 0.96 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr11_-_117747327 | 0.94 |
ENST00000584230.1 ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6 FXYD6-FXYD2 |
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr14_-_35344093 | 0.91 |
ENST00000382422.2 |
BAZ1A |
bromodomain adjacent to zinc finger domain, 1A |
| chr1_-_43424500 | 0.90 |
ENST00000415851.2 ENST00000426263.3 ENST00000372500.3 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
| chr19_-_18717627 | 0.89 |
ENST00000392386.3 |
CRLF1 |
cytokine receptor-like factor 1 |
| chr2_-_39348137 | 0.87 |
ENST00000426016.1 |
SOS1 |
son of sevenless homolog 1 (Drosophila) |
| chr7_+_29234028 | 0.87 |
ENST00000222792.6 |
CHN2 |
chimerin 2 |
| chr7_+_115850547 | 0.83 |
ENST00000358204.4 ENST00000455989.1 ENST00000537767.1 |
TES |
testis derived transcript (3 LIM domains) |
| chrX_-_39956656 | 0.83 |
ENST00000397354.3 ENST00000378444.4 |
BCOR |
BCL6 corepressor |
| chr15_+_50474385 | 0.81 |
ENST00000267842.5 |
SLC27A2 |
solute carrier family 27 (fatty acid transporter), member 2 |
| chrX_-_32173579 | 0.77 |
ENST00000359836.1 ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD |
dystrophin |
| chrX_+_46937745 | 0.77 |
ENST00000397180.1 ENST00000457380.1 ENST00000352078.4 |
RGN |
regucalcin |
| chrX_-_135962876 | 0.76 |
ENST00000431446.3 ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX |
RNA binding motif protein, X-linked |
| chr7_+_29234101 | 0.75 |
ENST00000435288.2 |
CHN2 |
chimerin 2 |
| chr3_-_57199397 | 0.75 |
ENST00000296318.7 |
IL17RD |
interleukin 17 receptor D |
| chr1_-_115053781 | 0.74 |
ENST00000358465.2 ENST00000369543.2 |
TRIM33 |
tripartite motif containing 33 |
| chr19_+_8274185 | 0.74 |
ENST00000558268.1 ENST00000558331.1 |
CERS4 |
ceramide synthase 4 |
| chr9_-_35650900 | 0.73 |
ENST00000259608.3 |
SIT1 |
signaling threshold regulating transmembrane adaptor 1 |
| chr21_-_46330545 | 0.73 |
ENST00000320216.6 ENST00000397852.1 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr1_+_244998602 | 0.72 |
ENST00000411948.2 |
COX20 |
COX20 cytochrome C oxidase assembly factor |
| chr17_+_72428218 | 0.72 |
ENST00000392628.2 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr19_+_8274204 | 0.71 |
ENST00000561053.1 ENST00000251363.5 ENST00000559450.1 ENST00000559336.1 |
CERS4 |
ceramide synthase 4 |
| chr20_+_55966444 | 0.70 |
ENST00000356208.5 ENST00000440234.2 |
RBM38 |
RNA binding motif protein 38 |
| chr6_-_89927151 | 0.69 |
ENST00000454853.2 |
GABRR1 |
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr15_+_43985725 | 0.69 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr8_-_70745575 | 0.69 |
ENST00000524945.1 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr3_-_47823298 | 0.67 |
ENST00000254480.5 |
SMARCC1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chr1_-_54304212 | 0.64 |
ENST00000540001.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr10_+_114710425 | 0.62 |
ENST00000352065.5 ENST00000369395.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr1_+_16085244 | 0.62 |
ENST00000400773.1 |
FBLIM1 |
filamin binding LIM protein 1 |
| chr15_+_41136216 | 0.61 |
ENST00000562057.1 ENST00000344051.4 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr14_-_74485960 | 0.60 |
ENST00000556242.1 ENST00000334696.6 |
ENTPD5 |
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr17_+_30813576 | 0.60 |
ENST00000313401.3 |
CDK5R1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr5_-_138725594 | 0.60 |
ENST00000302125.8 |
MZB1 |
marginal zone B and B1 cell-specific protein |
| chr5_-_138725560 | 0.60 |
ENST00000412103.2 ENST00000457570.2 |
MZB1 |
marginal zone B and B1 cell-specific protein |
| chr11_-_76155618 | 0.60 |
ENST00000530759.1 |
RP11-111M22.3 |
RP11-111M22.3 |
| chr6_+_32146131 | 0.60 |
ENST00000375094.3 |
RNF5 |
ring finger protein 5, E3 ubiquitin protein ligase |
| chr4_-_103266219 | 0.59 |
ENST00000394833.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chrX_-_110655391 | 0.59 |
ENST00000356915.2 ENST00000356220.3 |
DCX |
doublecortin |
| chr4_+_79567362 | 0.58 |
ENST00000512322.1 |
RP11-792D21.2 |
long intergenic non-protein coding RNA 1094 |
| chr1_-_54303949 | 0.58 |
ENST00000234725.8 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chrX_-_110655306 | 0.57 |
ENST00000371993.2 |
DCX |
doublecortin |
| chr6_+_64281906 | 0.57 |
ENST00000370651.3 |
PTP4A1 |
protein tyrosine phosphatase type IVA, member 1 |
| chr13_-_39612176 | 0.56 |
ENST00000352251.3 ENST00000350125.3 |
PROSER1 |
proline and serine rich 1 |
| chr11_-_76155700 | 0.55 |
ENST00000572035.1 |
RP11-111M22.3 |
RP11-111M22.3 |
| chr4_-_103266355 | 0.55 |
ENST00000424970.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr1_-_200992827 | 0.55 |
ENST00000332129.2 ENST00000422435.2 |
KIF21B |
kinesin family member 21B |
| chr17_-_39507064 | 0.55 |
ENST00000007735.3 |
KRT33A |
keratin 33A |
| chr10_+_114710211 | 0.55 |
ENST00000349937.2 ENST00000369397.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr7_+_73245193 | 0.54 |
ENST00000340958.2 |
CLDN4 |
claudin 4 |
| chr5_+_66124590 | 0.53 |
ENST00000490016.2 ENST00000403666.1 ENST00000450827.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr17_-_38020392 | 0.53 |
ENST00000346872.3 ENST00000439167.2 ENST00000377945.3 ENST00000394189.2 ENST00000377944.3 ENST00000377958.2 ENST00000535189.1 ENST00000377952.2 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
| chr11_+_77184416 | 0.53 |
ENST00000598970.1 |
DKFZP434E1119 |
DKFZP434E1119 |
| chr7_-_148725733 | 0.51 |
ENST00000286091.4 |
PDIA4 |
protein disulfide isomerase family A, member 4 |
| chr17_+_75447326 | 0.51 |
ENST00000591088.1 |
SEPT9 |
septin 9 |
| chr10_+_114710516 | 0.51 |
ENST00000542695.1 ENST00000346198.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr2_-_128568721 | 0.50 |
ENST00000322313.4 ENST00000393006.1 ENST00000409658.3 ENST00000436787.1 |
WDR33 |
WD repeat domain 33 |
| chr1_-_54303934 | 0.50 |
ENST00000537333.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr3_+_123813543 | 0.50 |
ENST00000360013.3 |
KALRN |
kalirin, RhoGEF kinase |
| chr17_+_79679299 | 0.49 |
ENST00000331531.5 |
SLC25A10 |
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr16_-_89007491 | 0.49 |
ENST00000327483.5 ENST00000564416.1 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr8_+_11660120 | 0.48 |
ENST00000220584.4 |
FDFT1 |
farnesyl-diphosphate farnesyltransferase 1 |
| chr4_+_79567314 | 0.48 |
ENST00000503539.1 ENST00000504675.1 |
RP11-792D21.2 |
long intergenic non-protein coding RNA 1094 |
| chr15_+_75074410 | 0.47 |
ENST00000439220.2 |
CSK |
c-src tyrosine kinase |
| chr16_+_19222479 | 0.46 |
ENST00000568433.1 |
SYT17 |
synaptotagmin XVII |
| chr8_+_11660227 | 0.46 |
ENST00000443614.2 ENST00000525900.1 |
FDFT1 |
farnesyl-diphosphate farnesyltransferase 1 |
| chr21_-_34143971 | 0.45 |
ENST00000290178.4 |
PAXBP1 |
PAX3 and PAX7 binding protein 1 |
| chr14_+_39644387 | 0.45 |
ENST00000553331.1 ENST00000216832.4 |
PNN |
pinin, desmosome associated protein |
| chr7_+_73868439 | 0.44 |
ENST00000424337.2 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
| chr4_-_109090106 | 0.44 |
ENST00000379951.2 |
LEF1 |
lymphoid enhancer-binding factor 1 |
| chr11_-_46615498 | 0.41 |
ENST00000533727.1 ENST00000534300.1 ENST00000528950.1 ENST00000526606.1 |
AMBRA1 |
autophagy/beclin-1 regulator 1 |
| chr3_+_141106643 | 0.41 |
ENST00000514251.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr12_-_48963829 | 0.40 |
ENST00000301046.2 ENST00000549817.1 |
LALBA |
lactalbumin, alpha- |
| chr2_-_100721923 | 0.40 |
ENST00000356421.2 |
AFF3 |
AF4/FMR2 family, member 3 |
| chr7_+_139528952 | 0.39 |
ENST00000416849.2 ENST00000436047.2 ENST00000414508.2 ENST00000448866.1 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
| chr3_-_167813132 | 0.39 |
ENST00000309027.4 |
GOLIM4 |
golgi integral membrane protein 4 |
| chr3_-_149375783 | 0.37 |
ENST00000467467.1 ENST00000460517.1 ENST00000360632.3 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr20_+_57875658 | 0.37 |
ENST00000371025.3 |
EDN3 |
endothelin 3 |
| chr7_+_128828713 | 0.36 |
ENST00000249373.3 |
SMO |
smoothened, frizzled family receptor |
| chr1_-_53793725 | 0.35 |
ENST00000371454.2 |
LRP8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr8_-_42234745 | 0.35 |
ENST00000220812.2 |
DKK4 |
dickkopf WNT signaling pathway inhibitor 4 |
| chr1_+_166808692 | 0.35 |
ENST00000367876.4 |
POGK |
pogo transposable element with KRAB domain |
| chr3_+_189349162 | 0.35 |
ENST00000264731.3 ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63 |
tumor protein p63 |
| chr16_-_15736881 | 0.34 |
ENST00000540441.2 |
KIAA0430 |
KIAA0430 |
| chr20_+_45947246 | 0.34 |
ENST00000599904.1 |
AL031666.2 |
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
| chr2_-_241725781 | 0.34 |
ENST00000428768.1 |
KIF1A |
kinesin family member 1A |
| chrX_+_100646190 | 0.33 |
ENST00000471855.1 |
RPL36A |
ribosomal protein L36a |
| chr5_-_114515734 | 0.33 |
ENST00000514154.1 ENST00000282369.3 |
TRIM36 |
tripartite motif containing 36 |
| chr16_-_4852915 | 0.32 |
ENST00000322048.7 |
ROGDI |
rogdi homolog (Drosophila) |
| chr4_-_120243545 | 0.32 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr21_+_30671690 | 0.31 |
ENST00000399921.1 |
BACH1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr1_-_109935819 | 0.31 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chr5_-_160973649 | 0.31 |
ENST00000393959.1 ENST00000517547.1 |
GABRB2 |
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr6_-_33548006 | 0.31 |
ENST00000374467.3 |
BAK1 |
BCL2-antagonist/killer 1 |
| chr18_+_29769978 | 0.31 |
ENST00000269202.6 ENST00000581447.1 |
MEP1B |
meprin A, beta |
| chr6_+_96025341 | 0.30 |
ENST00000369293.1 ENST00000358812.4 |
MANEA |
mannosidase, endo-alpha |
| chr15_-_89764929 | 0.30 |
ENST00000268125.5 |
RLBP1 |
retinaldehyde binding protein 1 |
| chr6_-_11779840 | 0.29 |
ENST00000506810.1 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr17_+_9066252 | 0.29 |
ENST00000436734.1 |
NTN1 |
netrin 1 |
| chr8_+_29953163 | 0.29 |
ENST00000518192.1 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
| chr6_+_108881012 | 0.29 |
ENST00000343882.6 |
FOXO3 |
forkhead box O3 |
| chr11_-_65381643 | 0.29 |
ENST00000309100.3 ENST00000529839.1 ENST00000526293.1 |
MAP3K11 |
mitogen-activated protein kinase kinase kinase 11 |
| chr12_+_109915179 | 0.28 |
ENST00000434735.2 |
UBE3B |
ubiquitin protein ligase E3B |
| chr2_+_32288725 | 0.28 |
ENST00000315285.3 |
SPAST |
spastin |
| chr15_-_79103757 | 0.28 |
ENST00000388820.4 |
ADAMTS7 |
ADAM metallopeptidase with thrombospondin type 1 motif, 7 |
| chr17_+_80416482 | 0.28 |
ENST00000309794.11 ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF |
nuclear prelamin A recognition factor |
| chr7_+_73703728 | 0.27 |
ENST00000361545.5 ENST00000223398.6 |
CLIP2 |
CAP-GLY domain containing linker protein 2 |
| chr3_+_52813932 | 0.27 |
ENST00000537050.1 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr1_-_177134024 | 0.26 |
ENST00000367654.3 |
ASTN1 |
astrotactin 1 |
| chr17_+_57297807 | 0.26 |
ENST00000284116.4 ENST00000581140.1 ENST00000581276.1 |
GDPD1 |
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr6_-_33547975 | 0.26 |
ENST00000442998.2 ENST00000360661.5 |
BAK1 |
BCL2-antagonist/killer 1 |
| chr20_+_57875758 | 0.26 |
ENST00000395654.3 |
EDN3 |
endothelin 3 |
| chr1_-_53793584 | 0.26 |
ENST00000354412.3 ENST00000347547.2 ENST00000306052.6 |
LRP8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr12_+_56324933 | 0.25 |
ENST00000549629.1 ENST00000555218.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr1_+_87794150 | 0.25 |
ENST00000370544.5 |
LMO4 |
LIM domain only 4 |
| chr16_+_2820912 | 0.25 |
ENST00000570539.1 |
SRRM2 |
serine/arginine repetitive matrix 2 |
| chr1_-_32801825 | 0.25 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chr4_+_95972822 | 0.24 |
ENST00000509540.1 ENST00000440890.2 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr4_+_170581213 | 0.24 |
ENST00000507875.1 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr2_+_169923577 | 0.24 |
ENST00000432060.2 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
| chr10_+_112631547 | 0.23 |
ENST00000280154.7 ENST00000393104.2 |
PDCD4 |
programmed cell death 4 (neoplastic transformation inhibitor) |
| chr6_-_11779174 | 0.23 |
ENST00000379413.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr8_-_17104356 | 0.23 |
ENST00000361272.4 ENST00000523917.1 |
CNOT7 |
CCR4-NOT transcription complex, subunit 7 |
| chr5_+_175288631 | 0.23 |
ENST00000509837.1 |
CPLX2 |
complexin 2 |
| chr1_-_6479963 | 0.22 |
ENST00000377836.4 ENST00000487437.1 ENST00000489730.1 ENST00000377834.4 |
HES2 |
hes family bHLH transcription factor 2 |
| chr20_+_44509857 | 0.22 |
ENST00000372523.1 ENST00000372520.1 |
ZSWIM1 |
zinc finger, SWIM-type containing 1 |
| chr5_+_112074029 | 0.21 |
ENST00000512211.2 |
APC |
adenomatous polyposis coli |
| chr8_-_33424636 | 0.21 |
ENST00000256257.1 |
RNF122 |
ring finger protein 122 |
| chr14_-_21516590 | 0.21 |
ENST00000555026.1 |
NDRG2 |
NDRG family member 2 |
| chr17_-_10372875 | 0.21 |
ENST00000255381.2 |
MYH4 |
myosin, heavy chain 4, skeletal muscle |
| chr21_-_34144157 | 0.21 |
ENST00000331923.4 |
PAXBP1 |
PAX3 and PAX7 binding protein 1 |
| chr2_+_169923504 | 0.20 |
ENST00000357546.2 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
| chr6_+_119215308 | 0.20 |
ENST00000229595.5 |
ASF1A |
anti-silencing function 1A histone chaperone |
| chr13_+_98086445 | 0.20 |
ENST00000245304.4 |
RAP2A |
RAP2A, member of RAS oncogene family |
| chr4_-_109089573 | 0.20 |
ENST00000265165.1 |
LEF1 |
lymphoid enhancer-binding factor 1 |
| chr3_+_152879985 | 0.20 |
ENST00000323534.2 |
RAP2B |
RAP2B, member of RAS oncogene family |
| chr5_+_149546334 | 0.19 |
ENST00000231656.8 |
CDX1 |
caudal type homeobox 1 |
| chrX_+_69488155 | 0.19 |
ENST00000374495.3 |
ARR3 |
arrestin 3, retinal (X-arrestin) |
| chr2_+_191273052 | 0.18 |
ENST00000417958.1 ENST00000432036.1 ENST00000392328.1 |
MFSD6 |
major facilitator superfamily domain containing 6 |
| chr4_-_69215467 | 0.18 |
ENST00000579690.1 |
YTHDC1 |
YTH domain containing 1 |
| chr3_+_183353356 | 0.18 |
ENST00000242810.6 ENST00000493074.1 ENST00000437402.1 ENST00000454495.2 ENST00000473045.1 ENST00000468101.1 ENST00000427201.2 ENST00000482138.1 ENST00000454652.2 |
KLHL24 |
kelch-like family member 24 |
| chr2_+_32288657 | 0.18 |
ENST00000345662.1 |
SPAST |
spastin |
| chr5_+_66300446 | 0.18 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr17_+_80416050 | 0.18 |
ENST00000579198.1 ENST00000390006.4 ENST00000580296.1 |
NARF |
nuclear prelamin A recognition factor |
| chr1_-_177133818 | 0.18 |
ENST00000424564.2 ENST00000361833.2 |
ASTN1 |
astrotactin 1 |
| chr19_+_41257084 | 0.17 |
ENST00000601393.1 |
SNRPA |
small nuclear ribonucleoprotein polypeptide A |
| chr12_+_15475462 | 0.17 |
ENST00000543886.1 ENST00000348962.2 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr16_-_18812746 | 0.17 |
ENST00000546206.2 ENST00000562819.1 ENST00000562234.2 ENST00000304414.7 ENST00000567078.2 |
ARL6IP1 RP11-1035H13.3 |
ADP-ribosylation factor-like 6 interacting protein 1 Uncharacterized protein |
| chr10_-_99052382 | 0.17 |
ENST00000453547.2 ENST00000316676.8 ENST00000358308.3 ENST00000466484.1 ENST00000358531.4 |
ARHGAP19-SLIT1 ARHGAP19 |
ARHGAP19-SLIT1 readthrough (NMD candidate) Rho GTPase activating protein 19 |
| chr6_+_134274322 | 0.16 |
ENST00000367871.1 ENST00000237264.4 |
TBPL1 |
TBP-like 1 |
| chr19_+_41256764 | 0.16 |
ENST00000243563.3 ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA |
small nuclear ribonucleoprotein polypeptide A |
| chr1_+_68150744 | 0.16 |
ENST00000370986.4 ENST00000370985.3 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
| chr19_+_11071652 | 0.16 |
ENST00000344626.4 ENST00000429416.3 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr12_+_31477250 | 0.15 |
ENST00000313737.4 |
AC024940.1 |
AC024940.1 |
| chr1_-_116383322 | 0.15 |
ENST00000429731.1 |
NHLH2 |
nescient helix loop helix 2 |
| chr2_-_166060552 | 0.15 |
ENST00000283254.7 ENST00000453007.1 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr10_-_104179682 | 0.15 |
ENST00000406432.1 |
PSD |
pleckstrin and Sec7 domain containing |
| chr4_-_186877502 | 0.15 |
ENST00000431902.1 ENST00000284776.7 ENST00000415274.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr9_-_36400920 | 0.14 |
ENST00000357058.3 ENST00000350199.4 |
RNF38 |
ring finger protein 38 |
| chr17_-_39526052 | 0.14 |
ENST00000251646.3 |
KRT33B |
keratin 33B |
| chr10_-_98945515 | 0.14 |
ENST00000371070.4 |
SLIT1 |
slit homolog 1 (Drosophila) |
| chr1_+_36348790 | 0.13 |
ENST00000373204.4 |
AGO1 |
argonaute RISC catalytic component 1 |
| chr6_-_11779403 | 0.13 |
ENST00000414691.3 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr14_+_75746781 | 0.13 |
ENST00000555347.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr14_-_71107921 | 0.12 |
ENST00000553982.1 ENST00000500016.1 |
CTD-2540L5.5 CTD-2540L5.6 |
CTD-2540L5.5 CTD-2540L5.6 |
| chr8_+_29952914 | 0.12 |
ENST00000321250.8 ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
| chr19_+_11071546 | 0.12 |
ENST00000358026.2 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chrX_+_100645812 | 0.12 |
ENST00000427805.2 ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A RPL36A-HNRNPH2 |
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr18_-_24445729 | 0.12 |
ENST00000383168.4 |
AQP4 |
aquaporin 4 |
| chrX_+_69488174 | 0.11 |
ENST00000480877.2 ENST00000307959.8 |
ARR3 |
arrestin 3, retinal (X-arrestin) |
| chr12_+_56324756 | 0.11 |
ENST00000331886.5 ENST00000555090.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr14_+_20923350 | 0.11 |
ENST00000555414.1 ENST00000216714.3 ENST00000553681.1 ENST00000557344.1 ENST00000398030.4 ENST00000557181.1 ENST00000555839.1 ENST00000553368.1 ENST00000556054.1 ENST00000557054.1 ENST00000557592.1 ENST00000557150.1 |
APEX1 |
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr4_-_69215699 | 0.11 |
ENST00000510746.1 ENST00000344157.4 ENST00000355665.3 |
YTHDC1 |
YTH domain containing 1 |
| chr1_-_1334685 | 0.10 |
ENST00000400809.3 ENST00000408918.4 |
CCNL2 |
cyclin L2 |
| chr7_-_148581251 | 0.10 |
ENST00000478654.1 ENST00000460911.1 ENST00000350995.2 |
EZH2 |
enhancer of zeste homolog 2 (Drosophila) |
| chr17_-_8263538 | 0.09 |
ENST00000535173.1 |
AC135178.1 |
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr10_+_76585303 | 0.09 |
ENST00000372725.1 |
KAT6B |
K(lysine) acetyltransferase 6B |
| chr10_-_91295304 | 0.08 |
ENST00000341233.4 ENST00000371790.4 |
SLC16A12 |
solute carrier family 16, member 12 |
| chrX_-_101726732 | 0.08 |
ENST00000457521.2 ENST00000412230.2 ENST00000453326.2 |
NXF2B TCP11X2 |
nuclear RNA export factor 2B t-complex 11 family, X-linked 2 |
| chr12_+_54402790 | 0.08 |
ENST00000040584.4 |
HOXC8 |
homeobox C8 |
| chr12_-_54694807 | 0.08 |
ENST00000435572.2 |
NFE2 |
nuclear factor, erythroid 2 |
| chr2_+_48796120 | 0.08 |
ENST00000394754.1 |
STON1-GTF2A1L |
STON1-GTF2A1L readthrough |
| chr7_-_148581360 | 0.08 |
ENST00000320356.2 ENST00000541220.1 ENST00000483967.1 ENST00000536783.1 |
EZH2 |
enhancer of zeste homolog 2 (Drosophila) |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.3 | 0.9 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.3 | 1.7 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 1.0 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.2 | 0.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 0.8 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 0.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.7 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 0.9 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.1 | 0.8 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.9 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 1.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.2 | GO:0044094 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.1 | 0.6 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.7 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.8 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 7.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 1.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.5 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.2 | GO:1990909 | catenin complex(GO:0016342) Wnt signalosome(GO:1990909) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 2.9 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.6 | 1.7 | GO:0051300 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.5 | 1.6 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.4 | 1.3 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.4 | 1.4 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.3 | 1.5 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.3 | 1.7 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.3 | 0.8 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.3 | 0.8 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.2 | 1.2 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.2 | 0.9 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 1.0 | GO:0044336 | embryonic genitalia morphogenesis(GO:0030538) canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.2 | 0.8 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.2 | 0.6 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.2 | 0.6 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.2 | 0.9 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.2 | 1.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 0.9 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.2 | 0.5 | GO:1902356 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.2 | 0.6 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.2 | 1.7 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.2 | 0.6 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 1.0 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.6 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.7 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 1.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 2.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.5 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 | 0.4 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.2 | GO:1905064 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 4.5 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 0.8 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.4 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.3 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.5 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.3 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.7 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.7 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.2 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.3 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.7 | GO:0000165 | MAPK cascade(GO:0000165) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.0 | 0.4 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.6 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.7 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.6 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.2 | GO:0060339 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.3 | GO:0051005 | endosome transport via multivesicular body sorting pathway(GO:0032509) negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.2 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 1.4 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.2 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.0 | 1.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.5 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0072560 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.2 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.0 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.1 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.0 | 0.5 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.5 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.3 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.7 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.4 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.4 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 1.8 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.5 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.2 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.3 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.0 | 0.2 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.7 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.4 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 4.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 4.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 4.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.9 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.0 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 2.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.1 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.5 | 1.6 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.3 | 0.9 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.3 | 2.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 1.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 0.9 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.2 | 1.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 0.6 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.2 | 0.6 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.2 | 0.9 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.2 | 0.5 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.2 | 0.6 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 0.4 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 2.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.6 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 3.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.7 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 0.4 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 1.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 1.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.3 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.3 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 5.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.8 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.5 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.6 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) primary miRNA binding(GO:0070878) |
| 0.0 | 0.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 2.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.5 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |