Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
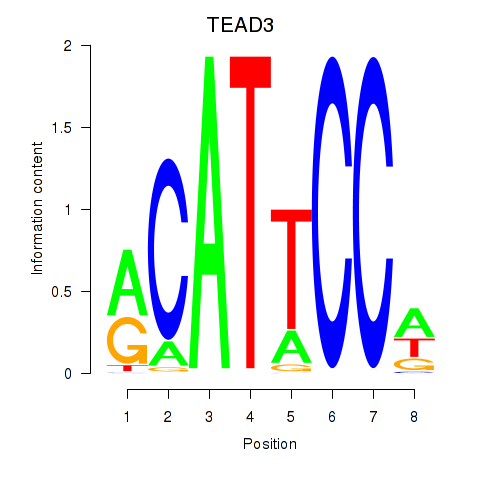
Results for TEAD3_TEAD1
Z-value: 4.78
Transcription factors associated with TEAD3_TEAD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TEAD3
|
ENSG00000007866.14 | TEAD3 |
|
TEAD1
|
ENSG00000187079.10 | TEAD1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TEAD1 | hg19_v2_chr11_+_12695944_12695989, hg19_v2_chr11_+_12696102_12696164, hg19_v2_chr11_+_12766583_12766592 | 0.94 | 1.1e-07 | Click! |
| TEAD3 | hg19_v2_chr6_-_35464817_35464894, hg19_v2_chr6_-_35464727_35464738 | 0.64 | 7.2e-03 | Click! |
Activity profile of TEAD3_TEAD1 motif
Sorted Z-values of TEAD3_TEAD1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TEAD3_TEAD1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_132272504 | 48.89 |
ENST00000367976.3 |
CTGF |
connective tissue growth factor |
| chr5_-_39425068 | 33.20 |
ENST00000515700.1 ENST00000339788.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr2_-_161264385 | 18.94 |
ENST00000409972.1 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr7_+_134464414 | 18.86 |
ENST00000361901.2 |
CALD1 |
caldesmon 1 |
| chr6_-_128841503 | 18.55 |
ENST00000368215.3 ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
| chr10_-_90712520 | 17.95 |
ENST00000224784.6 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr1_+_86046433 | 17.72 |
ENST00000451137.2 |
CYR61 |
cysteine-rich, angiogenic inducer, 61 |
| chr5_-_39425222 | 16.82 |
ENST00000320816.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr15_+_39873268 | 16.64 |
ENST00000397591.2 ENST00000260356.5 |
THBS1 |
thrombospondin 1 |
| chr5_-_39425290 | 16.38 |
ENST00000545653.1 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr1_-_95391315 | 16.12 |
ENST00000545882.1 ENST00000415017.1 |
CNN3 |
calponin 3, acidic |
| chr7_+_134464376 | 15.60 |
ENST00000454108.1 ENST00000361675.2 |
CALD1 |
caldesmon 1 |
| chr4_-_187644930 | 15.50 |
ENST00000441802.2 |
FAT1 |
FAT atypical cadherin 1 |
| chr4_+_184020398 | 15.27 |
ENST00000403733.3 ENST00000378925.3 |
WWC2 |
WW and C2 domain containing 2 |
| chr15_+_63334831 | 14.09 |
ENST00000288398.6 ENST00000358278.3 ENST00000560445.1 ENST00000403994.3 ENST00000357980.4 ENST00000559556.1 ENST00000559397.1 ENST00000267996.7 ENST00000560970.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr2_-_216257849 | 13.74 |
ENST00000456923.1 |
FN1 |
fibronectin 1 |
| chr7_+_134430212 | 13.52 |
ENST00000436461.2 |
CALD1 |
caldesmon 1 |
| chr16_+_30387141 | 11.98 |
ENST00000566955.1 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr10_-_17659234 | 11.86 |
ENST00000466335.1 |
PTPLA |
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chrX_-_10588459 | 11.75 |
ENST00000380782.2 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr3_-_134093275 | 11.58 |
ENST00000513145.1 ENST00000422605.2 |
AMOTL2 |
angiomotin like 2 |
| chr11_-_111781454 | 11.58 |
ENST00000533280.1 |
CRYAB |
crystallin, alpha B |
| chr7_+_116165754 | 11.47 |
ENST00000405348.1 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
| chr5_-_55290773 | 11.01 |
ENST00000502326.3 ENST00000381298.2 |
IL6ST |
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chr3_-_134093395 | 10.61 |
ENST00000249883.5 |
AMOTL2 |
angiomotin like 2 |
| chr4_+_169552748 | 10.35 |
ENST00000504519.1 ENST00000512127.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr1_-_1293904 | 10.10 |
ENST00000309212.6 ENST00000342753.4 ENST00000445648.2 |
MXRA8 |
matrix-remodelling associated 8 |
| chr7_+_116165038 | 9.93 |
ENST00000393470.1 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
| chr1_-_201346761 | 9.69 |
ENST00000455702.1 ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2 |
troponin T type 2 (cardiac) |
| chr16_-_122619 | 9.53 |
ENST00000262316.6 |
RHBDF1 |
rhomboid 5 homolog 1 (Drosophila) |
| chr1_-_94050668 | 8.90 |
ENST00000539242.1 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr10_+_123872483 | 8.83 |
ENST00000369001.1 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr4_-_186733363 | 8.70 |
ENST00000393523.2 ENST00000393528.3 ENST00000449407.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr11_+_114166536 | 8.51 |
ENST00000299964.3 |
NNMT |
nicotinamide N-methyltransferase |
| chr3_-_114343039 | 8.47 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr16_+_30386098 | 7.78 |
ENST00000322861.7 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr17_-_40575535 | 7.67 |
ENST00000357037.5 |
PTRF |
polymerase I and transcript release factor |
| chr8_+_70378852 | 7.66 |
ENST00000525061.1 ENST00000458141.2 ENST00000260128.4 |
SULF1 |
sulfatase 1 |
| chr11_-_47470591 | 7.63 |
ENST00000524487.1 |
RAPSN |
receptor-associated protein of the synapse |
| chr7_+_128470431 | 7.48 |
ENST00000325888.8 ENST00000346177.6 |
FLNC |
filamin C, gamma |
| chr10_-_99447024 | 7.38 |
ENST00000370626.3 |
AVPI1 |
arginine vasopressin-induced 1 |
| chr2_-_190044480 | 7.34 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr11_-_47470703 | 7.26 |
ENST00000298854.2 |
RAPSN |
receptor-associated protein of the synapse |
| chr10_-_33247124 | 7.20 |
ENST00000414670.1 ENST00000302278.3 ENST00000374956.4 ENST00000488494.1 ENST00000417122.2 ENST00000474568.1 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
| chr3_-_185538849 | 7.16 |
ENST00000421047.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr20_+_35169885 | 6.96 |
ENST00000279022.2 ENST00000346786.2 |
MYL9 |
myosin, light chain 9, regulatory |
| chr11_-_47470682 | 6.85 |
ENST00000529341.1 ENST00000352508.3 |
RAPSN |
receptor-associated protein of the synapse |
| chr2_-_20212422 | 6.81 |
ENST00000421259.2 ENST00000407540.3 |
MATN3 |
matrilin 3 |
| chr15_+_33010175 | 6.76 |
ENST00000300177.4 ENST00000560677.1 ENST00000560830.1 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chr11_-_111781610 | 6.30 |
ENST00000525823.1 |
CRYAB |
crystallin, alpha B |
| chr12_-_8815299 | 6.25 |
ENST00000535336.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr11_-_111782696 | 6.22 |
ENST00000227251.3 ENST00000526180.1 |
CRYAB |
crystallin, alpha B |
| chr1_+_114522049 | 6.11 |
ENST00000369551.1 ENST00000320334.4 |
OLFML3 |
olfactomedin-like 3 |
| chr12_-_8815215 | 5.97 |
ENST00000544889.1 ENST00000543369.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr12_-_7245125 | 5.92 |
ENST00000542285.1 ENST00000540610.1 |
C1R |
complement component 1, r subcomponent |
| chr1_-_16482554 | 5.80 |
ENST00000358432.5 |
EPHA2 |
EPH receptor A2 |
| chrX_-_6146876 | 5.53 |
ENST00000381095.3 |
NLGN4X |
neuroligin 4, X-linked |
| chr11_-_8832182 | 5.45 |
ENST00000527510.1 ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5 |
suppression of tumorigenicity 5 |
| chr6_+_30850697 | 5.30 |
ENST00000509639.1 ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr12_-_54813229 | 5.25 |
ENST00000293379.4 |
ITGA5 |
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr12_-_8815404 | 5.20 |
ENST00000359478.2 ENST00000396549.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr11_-_111781554 | 5.12 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chr11_-_111782484 | 5.09 |
ENST00000533971.1 |
CRYAB |
crystallin, alpha B |
| chrX_-_10851762 | 5.09 |
ENST00000380785.1 ENST00000380787.1 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr3_+_147127142 | 4.91 |
ENST00000282928.4 |
ZIC1 |
Zic family member 1 |
| chr2_-_192711968 | 4.72 |
ENST00000304141.4 |
SDPR |
serum deprivation response |
| chr15_+_96869165 | 4.57 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr9_-_13175823 | 4.51 |
ENST00000545857.1 |
MPDZ |
multiple PDZ domain protein |
| chr15_-_60690163 | 4.39 |
ENST00000558998.1 ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2 |
annexin A2 |
| chr6_+_83072923 | 4.38 |
ENST00000535040.1 |
TPBG |
trophoblast glycoprotein |
| chr21_+_30502806 | 4.27 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr2_-_211179883 | 4.23 |
ENST00000352451.3 |
MYL1 |
myosin, light chain 1, alkali; skeletal, fast |
| chr11_-_14379997 | 4.14 |
ENST00000526063.1 ENST00000532814.1 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
| chr6_+_83073334 | 4.13 |
ENST00000369750.3 |
TPBG |
trophoblast glycoprotein |
| chr19_-_50143452 | 4.10 |
ENST00000246792.3 |
RRAS |
related RAS viral (r-ras) oncogene homolog |
| chr19_-_14606900 | 3.98 |
ENST00000393029.3 ENST00000393028.1 ENST00000393033.4 ENST00000345425.2 ENST00000586027.1 ENST00000591349.1 ENST00000587210.1 |
GIPC1 |
GIPC PDZ domain containing family, member 1 |
| chr19_+_15218180 | 3.95 |
ENST00000342784.2 ENST00000597977.1 ENST00000600440.1 |
SYDE1 |
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr3_-_127541194 | 3.92 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr15_-_48937982 | 3.85 |
ENST00000316623.5 |
FBN1 |
fibrillin 1 |
| chr15_+_63354769 | 3.84 |
ENST00000558910.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr9_+_131314859 | 3.75 |
ENST00000358161.5 ENST00000372731.4 ENST00000372739.3 |
SPTAN1 |
spectrin, alpha, non-erythrocytic 1 |
| chr9_-_16727978 | 3.72 |
ENST00000418777.1 ENST00000468187.2 |
BNC2 |
basonuclin 2 |
| chr13_-_33760216 | 3.60 |
ENST00000255486.4 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr11_+_111782934 | 3.43 |
ENST00000304298.3 |
HSPB2 |
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr17_-_39677971 | 3.39 |
ENST00000393976.2 |
KRT15 |
keratin 15 |
| chr12_-_104443890 | 3.39 |
ENST00000547583.1 ENST00000360814.4 ENST00000546851.1 |
GLT8D2 |
glycosyltransferase 8 domain containing 2 |
| chrX_+_99899180 | 3.38 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr1_-_229569834 | 3.36 |
ENST00000366684.3 ENST00000366683.2 |
ACTA1 |
actin, alpha 1, skeletal muscle |
| chr1_-_94079648 | 3.33 |
ENST00000370247.3 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr18_-_25616519 | 3.33 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chr4_-_186877806 | 3.31 |
ENST00000355634.5 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr4_+_41614909 | 3.22 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr9_+_112542591 | 3.21 |
ENST00000483909.1 ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2 PALM2-AKAP2 AKAP2 |
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr1_-_201438282 | 3.20 |
ENST00000367311.3 ENST00000367309.1 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
| chr19_-_51512804 | 3.18 |
ENST00000594211.1 ENST00000376832.4 |
KLK9 |
kallikrein-related peptidase 9 |
| chr9_+_139873264 | 3.16 |
ENST00000446677.1 |
PTGDS |
prostaglandin D2 synthase 21kDa (brain) |
| chr9_-_139891165 | 3.11 |
ENST00000494426.1 |
CLIC3 |
chloride intracellular channel 3 |
| chr8_+_1772132 | 3.05 |
ENST00000349830.3 ENST00000520359.1 ENST00000518288.1 ENST00000398560.1 |
ARHGEF10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr8_+_97597148 | 3.02 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr16_+_14802801 | 3.01 |
ENST00000526520.1 ENST00000531598.2 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr4_-_186877502 | 3.00 |
ENST00000431902.1 ENST00000284776.7 ENST00000415274.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr19_+_39138320 | 2.93 |
ENST00000424234.2 ENST00000390009.3 ENST00000589528.1 |
ACTN4 |
actinin, alpha 4 |
| chr3_-_149375783 | 2.84 |
ENST00000467467.1 ENST00000460517.1 ENST00000360632.3 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr1_+_196621002 | 2.83 |
ENST00000367429.4 ENST00000439155.2 |
CFH |
complement factor H |
| chr6_+_123110302 | 2.75 |
ENST00000368440.4 |
SMPDL3A |
sphingomyelin phosphodiesterase, acid-like 3A |
| chr5_+_36608422 | 2.75 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr6_-_35464727 | 2.73 |
ENST00000402886.3 |
TEAD3 |
TEA domain family member 3 |
| chr9_+_112542572 | 2.72 |
ENST00000374530.3 |
PALM2-AKAP2 |
PALM2-AKAP2 readthrough |
| chr8_-_119124045 | 2.72 |
ENST00000378204.2 |
EXT1 |
exostosin glycosyltransferase 1 |
| chr3_-_52486841 | 2.71 |
ENST00000496590.1 |
TNNC1 |
troponin C type 1 (slow) |
| chr2_+_170366203 | 2.70 |
ENST00000284669.1 |
KLHL41 |
kelch-like family member 41 |
| chr11_+_101981169 | 2.70 |
ENST00000526343.1 ENST00000282441.5 ENST00000537274.1 ENST00000345877.2 |
YAP1 |
Yes-associated protein 1 |
| chr10_-_92681033 | 2.68 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr11_+_66824276 | 2.68 |
ENST00000308831.2 |
RHOD |
ras homolog family member D |
| chr10_-_33625154 | 2.66 |
ENST00000265371.4 |
NRP1 |
neuropilin 1 |
| chr7_-_150946015 | 2.66 |
ENST00000262188.8 |
SMARCD3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr2_-_217560248 | 2.64 |
ENST00000233813.4 |
IGFBP5 |
insulin-like growth factor binding protein 5 |
| chr8_-_125740514 | 2.63 |
ENST00000325064.5 ENST00000518547.1 |
MTSS1 |
metastasis suppressor 1 |
| chr6_+_123110465 | 2.61 |
ENST00000539041.1 |
SMPDL3A |
sphingomyelin phosphodiesterase, acid-like 3A |
| chr13_+_32605437 | 2.57 |
ENST00000380250.3 |
FRY |
furry homolog (Drosophila) |
| chrX_-_41449204 | 2.57 |
ENST00000378179.3 |
CASK |
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr9_+_103204553 | 2.50 |
ENST00000502978.1 ENST00000334943.6 |
MSANTD3-TMEFF1 TMEFF1 |
MSANTD3-TMEFF1 readthrough transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr6_-_35464817 | 2.45 |
ENST00000338863.7 |
TEAD3 |
TEA domain family member 3 |
| chr18_+_7754957 | 2.43 |
ENST00000400053.4 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
| chr16_-_15474904 | 2.39 |
ENST00000534094.1 |
NPIPA5 |
nuclear pore complex interacting protein family, member A5 |
| chr9_-_14314566 | 2.37 |
ENST00000397579.2 |
NFIB |
nuclear factor I/B |
| chr4_-_7873981 | 2.27 |
ENST00000360265.4 |
AFAP1 |
actin filament associated protein 1 |
| chr9_-_14314518 | 2.26 |
ENST00000397581.2 |
NFIB |
nuclear factor I/B |
| chr10_+_123923205 | 2.26 |
ENST00000369004.3 ENST00000260733.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr11_-_64013663 | 2.25 |
ENST00000392210.2 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr19_+_676385 | 2.24 |
ENST00000166139.4 |
FSTL3 |
follistatin-like 3 (secreted glycoprotein) |
| chr6_+_148663729 | 2.20 |
ENST00000367467.3 |
SASH1 |
SAM and SH3 domain containing 1 |
| chr4_-_186732048 | 2.20 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr5_+_140868717 | 2.09 |
ENST00000252087.1 |
PCDHGC5 |
protocadherin gamma subfamily C, 5 |
| chr17_+_37856299 | 2.03 |
ENST00000269571.5 |
ERBB2 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr15_+_43809797 | 1.99 |
ENST00000399453.1 ENST00000300231.5 |
MAP1A |
microtubule-associated protein 1A |
| chr11_-_62521614 | 1.97 |
ENST00000527994.1 ENST00000394807.3 |
ZBTB3 |
zinc finger and BTB domain containing 3 |
| chr17_-_10325261 | 1.97 |
ENST00000403437.2 |
MYH8 |
myosin, heavy chain 8, skeletal muscle, perinatal |
| chr11_-_76381029 | 1.97 |
ENST00000407242.2 ENST00000421973.1 |
LRRC32 |
leucine rich repeat containing 32 |
| chr18_+_3247779 | 1.95 |
ENST00000578611.1 ENST00000583449.1 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr1_+_196621156 | 1.95 |
ENST00000359637.2 |
CFH |
complement factor H |
| chr8_-_108510224 | 1.94 |
ENST00000517746.1 ENST00000297450.3 |
ANGPT1 |
angiopoietin 1 |
| chr12_-_8815477 | 1.93 |
ENST00000433590.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr15_+_62853562 | 1.92 |
ENST00000561311.1 |
TLN2 |
talin 2 |
| chr19_+_39138271 | 1.76 |
ENST00000252699.2 |
ACTN4 |
actinin, alpha 4 |
| chr18_+_3247413 | 1.74 |
ENST00000579226.1 ENST00000217652.3 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr7_+_100464760 | 1.74 |
ENST00000200457.4 |
TRIP6 |
thyroid hormone receptor interactor 6 |
| chr8_+_94929969 | 1.73 |
ENST00000517764.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr15_-_42565606 | 1.70 |
ENST00000307216.6 ENST00000448392.1 |
TMEM87A |
transmembrane protein 87A |
| chr1_-_109935819 | 1.68 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chrY_+_16634483 | 1.64 |
ENST00000382872.1 |
NLGN4Y |
neuroligin 4, Y-linked |
| chr11_+_66824346 | 1.61 |
ENST00000532559.1 |
RHOD |
ras homolog family member D |
| chr1_-_227506158 | 1.61 |
ENST00000366769.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr4_+_166300084 | 1.59 |
ENST00000402744.4 |
CPE |
carboxypeptidase E |
| chrX_-_23761317 | 1.57 |
ENST00000492081.1 ENST00000379303.5 ENST00000336430.7 |
ACOT9 |
acyl-CoA thioesterase 9 |
| chr14_-_23904861 | 1.56 |
ENST00000355349.3 |
MYH7 |
myosin, heavy chain 7, cardiac muscle, beta |
| chr2_+_102759199 | 1.56 |
ENST00000409288.1 ENST00000410023.1 |
IL1R1 |
interleukin 1 receptor, type I |
| chr6_+_143999072 | 1.51 |
ENST00000440869.2 ENST00000367582.3 ENST00000451827.2 |
PHACTR2 |
phosphatase and actin regulator 2 |
| chr20_-_14318248 | 1.49 |
ENST00000378053.3 ENST00000341420.4 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
| chr10_-_75410771 | 1.49 |
ENST00000372873.4 |
SYNPO2L |
synaptopodin 2-like |
| chr13_-_67804445 | 1.47 |
ENST00000456367.1 ENST00000377861.3 ENST00000544246.1 |
PCDH9 |
protocadherin 9 |
| chr17_+_37856253 | 1.47 |
ENST00000540147.1 ENST00000584450.1 |
ERBB2 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr7_-_27219849 | 1.47 |
ENST00000396344.4 |
HOXA10 |
homeobox A10 |
| chr17_-_7297833 | 1.46 |
ENST00000571802.1 ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr1_-_205290865 | 1.45 |
ENST00000367157.3 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
| chr12_+_3068466 | 1.44 |
ENST00000358409.2 |
TEAD4 |
TEA domain family member 4 |
| chr17_+_43224684 | 1.42 |
ENST00000332499.2 |
HEXIM1 |
hexamethylene bis-acetamide inducible 1 |
| chr6_-_3157760 | 1.42 |
ENST00000333628.3 |
TUBB2A |
tubulin, beta 2A class IIa |
| chr20_+_3451650 | 1.42 |
ENST00000262919.5 |
ATRN |
attractin |
| chr12_-_47219733 | 1.41 |
ENST00000547477.1 ENST00000447411.1 ENST00000266579.4 |
SLC38A4 |
solute carrier family 38, member 4 |
| chr18_+_55862622 | 1.36 |
ENST00000456173.2 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr20_+_44044717 | 1.33 |
ENST00000279036.6 ENST00000279035.9 ENST00000372689.5 ENST00000545755.1 ENST00000341555.5 ENST00000535404.1 ENST00000543458.2 ENST00000432270.1 |
PIGT |
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr16_+_1578674 | 1.31 |
ENST00000253934.5 |
TMEM204 |
transmembrane protein 204 |
| chr14_+_74035763 | 1.31 |
ENST00000238651.5 |
ACOT2 |
acyl-CoA thioesterase 2 |
| chr14_+_32546485 | 1.30 |
ENST00000345122.3 ENST00000432921.1 ENST00000433497.1 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr12_+_27677085 | 1.30 |
ENST00000545334.1 ENST00000540114.1 ENST00000537927.1 ENST00000318304.8 ENST00000535047.1 ENST00000542629.1 ENST00000228425.6 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
| chr6_-_152489484 | 1.30 |
ENST00000354674.4 ENST00000539504.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr1_-_209825674 | 1.30 |
ENST00000367030.3 ENST00000356082.4 |
LAMB3 |
laminin, beta 3 |
| chrX_+_153046456 | 1.28 |
ENST00000393786.3 ENST00000370104.1 ENST00000370108.3 ENST00000370101.3 ENST00000430541.1 ENST00000370100.1 |
SRPK3 |
SRSF protein kinase 3 |
| chr11_-_64013288 | 1.27 |
ENST00000542235.1 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr11_+_46299199 | 1.24 |
ENST00000529193.1 ENST00000288400.3 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
| chr12_+_3068544 | 1.22 |
ENST00000540314.1 ENST00000536826.1 ENST00000359864.2 |
TEAD4 |
TEA domain family member 4 |
| chr17_-_79105734 | 1.21 |
ENST00000417379.1 |
AATK |
apoptosis-associated tyrosine kinase |
| chr16_-_21436459 | 1.20 |
ENST00000448012.2 ENST00000504841.2 ENST00000419180.2 |
NPIPB3 |
nuclear pore complex interacting protein family, member B3 |
| chr17_+_37856214 | 1.19 |
ENST00000445658.2 |
ERBB2 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr5_-_95158644 | 1.18 |
ENST00000237858.6 |
GLRX |
glutaredoxin (thioltransferase) |
| chr6_-_52926539 | 1.17 |
ENST00000350082.5 ENST00000356971.3 |
ICK |
intestinal cell (MAK-like) kinase |
| chr9_-_23826298 | 1.17 |
ENST00000380117.1 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr16_+_30907927 | 1.15 |
ENST00000279804.2 ENST00000395019.3 |
CTF1 |
cardiotrophin 1 |
| chr1_+_178310581 | 1.10 |
ENST00000462775.1 |
RASAL2 |
RAS protein activator like 2 |
| chr11_+_7506713 | 1.10 |
ENST00000329293.3 ENST00000534244.1 |
OLFML1 |
olfactomedin-like 1 |
| chr12_-_111358372 | 1.09 |
ENST00000548438.1 ENST00000228841.8 |
MYL2 |
myosin, light chain 2, regulatory, cardiac, slow |
| chr17_+_7348658 | 1.08 |
ENST00000570557.1 ENST00000536404.2 ENST00000576360.1 |
CHRNB1 |
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr15_+_41245160 | 1.08 |
ENST00000444189.2 ENST00000446533.3 |
CHAC1 |
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr5_-_176923846 | 1.08 |
ENST00000506537.1 |
PDLIM7 |
PDZ and LIM domain 7 (enigma) |
| chr3_-_47950745 | 1.07 |
ENST00000429422.1 |
MAP4 |
microtubule-associated protein 4 |
| chr8_-_38325219 | 1.07 |
ENST00000533668.1 ENST00000413133.2 ENST00000397108.4 ENST00000526742.1 ENST00000525001.1 ENST00000425967.3 ENST00000529552.1 ENST00000397113.2 |
FGFR1 |
fibroblast growth factor receptor 1 |
| chr5_-_95158375 | 1.06 |
ENST00000512469.2 ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX |
glutaredoxin (thioltransferase) |
| chr12_-_120687948 | 1.05 |
ENST00000458477.2 |
PXN |
paxillin |
| chr12_+_120933859 | 1.05 |
ENST00000242577.6 ENST00000548214.1 ENST00000392508.2 |
DYNLL1 |
dynein, light chain, LC8-type 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.6 | 66.4 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 16.3 | 48.9 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 7.2 | 21.7 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 7.1 | 21.4 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 5.5 | 16.6 | GO:0002605 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) |
| 4.6 | 13.7 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 3.6 | 21.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 3.3 | 13.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 3.1 | 12.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 2.9 | 5.8 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 2.4 | 7.2 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 2.3 | 6.8 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 2.2 | 17.9 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 2.0 | 17.7 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) chondroblast differentiation(GO:0060591) |
| 1.9 | 34.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 1.8 | 7.3 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 1.7 | 3.3 | GO:1901631 | positive regulation of presynaptic membrane organization(GO:1901631) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 1.6 | 4.7 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 1.5 | 21.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 1.5 | 4.4 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 1.3 | 16.8 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 1.3 | 7.7 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 1.1 | 4.6 | GO:0009956 | radial pattern formation(GO:0009956) |
| 1.1 | 7.7 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 1.0 | 3.9 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.9 | 4.7 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.9 | 2.6 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.9 | 7.8 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.9 | 5.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.8 | 5.4 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.8 | 3.8 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.8 | 16.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.7 | 0.7 | GO:0072190 | ureter urothelium development(GO:0072190) ureter morphogenesis(GO:0072197) |
| 0.7 | 30.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.7 | 3.6 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.7 | 8.5 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.7 | 2.7 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.6 | 0.6 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.6 | 5.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.6 | 12.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.6 | 10.1 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.6 | 3.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.6 | 18.8 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.6 | 2.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.5 | 5.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.5 | 2.7 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.5 | 1.6 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.5 | 1.5 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.5 | 17.2 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.5 | 4.8 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.5 | 1.9 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.5 | 10.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.4 | 2.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.4 | 2.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.4 | 3.0 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.4 | 3.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.4 | 2.7 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.4 | 1.9 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.4 | 1.1 | GO:0007037 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.3 | 1.7 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.3 | 1.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.3 | 1.0 | GO:0072275 | branchiomeric skeletal muscle development(GO:0014707) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.3 | 2.6 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.3 | 1.6 | GO:0030070 | insulin processing(GO:0030070) |
| 0.3 | 1.2 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.3 | 2.7 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 0.3 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.3 | 1.0 | GO:1904457 | glucosylceramide catabolic process(GO:0006680) termination of signal transduction(GO:0023021) beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.2 | 1.0 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.2 | 0.7 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.2 | 1.8 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 2.0 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.2 | 0.4 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 | 0.4 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.2 | 6.5 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.2 | 3.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 4.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 0.7 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.2 | 2.7 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.2 | 0.2 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.2 | 4.2 | GO:0007628 | adult walking behavior(GO:0007628) |
| 0.2 | 1.7 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 | 2.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.2 | 1.5 | GO:0060973 | cell migration involved in heart development(GO:0060973) |
| 0.2 | 0.5 | GO:1904386 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.2 | 2.3 | GO:0043586 | tongue development(GO:0043586) |
| 0.2 | 2.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.2 | 4.0 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 75.7 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 1.4 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 | 1.3 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 1.4 | GO:0007017 | microtubule-based process(GO:0007017) |
| 0.1 | 6.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.2 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.1 | 2.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.4 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.1 | 8.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.3 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 16.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.5 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.1 | 2.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 3.4 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 | 1.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.4 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.3 | GO:0045112 | integrin biosynthetic process(GO:0045112) regulation of integrin biosynthetic process(GO:0045113) positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 0.3 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.1 | 0.6 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 0.3 | GO:0098743 | cell aggregation(GO:0098743) |
| 0.1 | 1.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.3 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.5 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 | 0.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 9.9 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 0.2 | GO:0006738 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.1 | 0.8 | GO:1905247 | positive regulation of beta-amyloid clearance(GO:1900223) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.1 | 0.2 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 0.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 3.2 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 0.5 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 4.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 1.5 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 1.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.2 | GO:0039650 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.1 | 7.0 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.1 | 6.8 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.1 | 0.4 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 | 1.3 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.1 | 1.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.8 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 1.0 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.6 | GO:0030220 | platelet formation(GO:0030220) |
| 0.1 | 0.7 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.1 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.1 | 0.6 | GO:0006702 | androgen biosynthetic process(GO:0006702) |
| 0.1 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 2.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 5.9 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 5.9 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 0.0 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 2.2 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 5.6 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 3.5 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) cellular amide catabolic process(GO:0043605) |
| 0.0 | 2.0 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.2 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.0 | 0.1 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.0 | 2.3 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 3.4 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 2.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.3 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.1 | GO:0061046 | regulation of branching involved in lung morphogenesis(GO:0061046) |
| 0.0 | 1.7 | GO:0016032 | viral process(GO:0016032) multi-organism cellular process(GO:0044764) |
| 0.0 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.4 | GO:1903393 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.8 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.3 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 7.0 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 1.3 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.3 | GO:0051931 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.0 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.3 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.9 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.6 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 21.4 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 5.5 | 16.6 | GO:0070052 | collagen V binding(GO:0070052) |
| 4.1 | 12.3 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 3.1 | 67.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 2.5 | 61.0 | GO:0001968 | fibronectin binding(GO:0001968) |
| 2.1 | 8.5 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 2.1 | 58.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 1.9 | 7.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.7 | 12.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 1.1 | 21.7 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 1.1 | 34.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.1 | 5.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.0 | 18.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.9 | 10.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.9 | 2.7 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.8 | 20.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.8 | 11.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.8 | 3.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.7 | 2.7 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.7 | 65.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.6 | 1.3 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.5 | 5.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.5 | 15.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.5 | 3.6 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.4 | 1.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.4 | 4.7 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.4 | 2.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 1.6 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.3 | 2.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.3 | 30.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.3 | 3.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.3 | 1.6 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.3 | 5.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.3 | 3.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 2.7 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.3 | 1.7 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 5.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.3 | 13.7 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.3 | 2.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 1.0 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.2 | 1.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 2.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.2 | 0.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.2 | 3.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 4.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 7.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 0.6 | GO:0004769 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 0.2 | 0.8 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.2 | 0.7 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 0.8 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 14.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 0.7 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.5 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 4.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 11.4 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 5.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 5.9 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 1.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.2 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.1 | 2.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 11.4 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 1.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 1.5 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 6.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 10.2 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.1 | 2.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 1.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.2 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 3.0 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 9.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 3.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.1 | 4.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 5.7 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.1 | 0.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 4.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.4 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.1 | 1.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 1.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 3.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.0 | 1.0 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 1.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.3 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 11.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 2.4 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0015403 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 3.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 5.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 1.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.7 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 0.2 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 1.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.7 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 4.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 3.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 3.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 140.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 1.3 | 35.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.6 | 32.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.3 | 6.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.3 | 18.0 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.2 | 19.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.2 | 15.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 5.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 2.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 2.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 2.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.2 | 14.9 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.2 | 12.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.2 | 7.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 7.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 6.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 7.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 4.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 3.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.6 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 1.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 8.4 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 1.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 9.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.9 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.1 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 1.2 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.9 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 8.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.8 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.7 | 66.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 1.7 | 64.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 1.4 | 91.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.9 | 22.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.9 | 24.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.5 | 14.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.5 | 11.8 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.4 | 6.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.3 | 8.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.3 | 15.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 1.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.2 | 3.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.2 | 5.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 4.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 14.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 7.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 3.0 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 3.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.8 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 2.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 7.3 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 1.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 2.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 1.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 3.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 5.6 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 4.3 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 2.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 5.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 1.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 66.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 4.5 | 18.0 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 3.2 | 47.3 | GO:0030478 | actin cap(GO:0030478) |
| 2.8 | 45.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 2.8 | 11.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 2.4 | 7.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.4 | 7.2 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 2.2 | 29.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.8 | 21.7 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 1.4 | 19.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 1.4 | 2.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 1.3 | 5.3 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 1.1 | 17.9 | GO:0032059 | bleb(GO:0032059) |
| 1.0 | 22.9 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.9 | 4.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.8 | 48.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.8 | 28.9 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.6 | 5.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.5 | 2.7 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.4 | 1.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.3 | 1.7 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.3 | 3.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.3 | 2.6 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.3 | 7.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.3 | 4.6 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.3 | 17.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 2.7 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 10.4 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 18.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 0.7 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 1.3 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 18.6 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.2 | 2.6 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.2 | 5.9 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 2.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 5.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 15.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 0.3 | GO:0071437 | invadopodium(GO:0071437) invadopodium membrane(GO:0071438) |
| 0.1 | 0.8 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.1 | 0.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.1 | 18.4 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 1.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 5.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 12.5 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 1.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 2.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 26.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.2 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 1.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.6 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 6.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 3.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 4.4 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 6.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 6.3 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 1.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 2.2 | GO:0005875 | microtubule associated complex(GO:0005875) |
| 0.0 | 0.5 | GO:0036379 | myofilament(GO:0036379) |
| 0.0 | 1.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.9 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.2 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) ripoptosome(GO:0097342) |
| 0.0 | 5.0 | GO:0010008 | endosome membrane(GO:0010008) |