Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
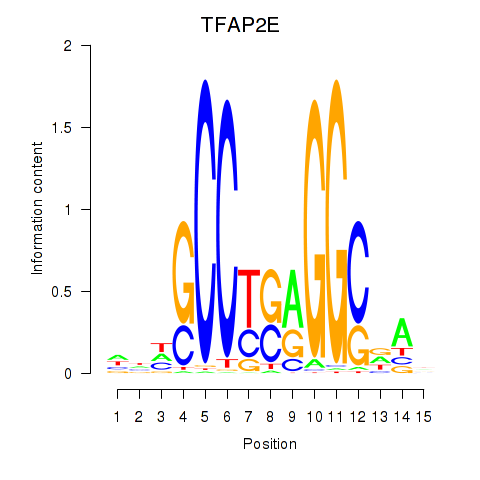
Results for TFAP2E
Z-value: 0.69
Transcription factors associated with TFAP2E
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2E
|
ENSG00000116819.6 | TFAP2E |
Activity profile of TFAP2E motif
Sorted Z-values of TFAP2E motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TFAP2E
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_116700614 | 2.29 |
ENST00000375345.1 |
APOC3 |
apolipoprotein C-III |
| chr11_+_116700600 | 2.28 |
ENST00000227667.3 |
APOC3 |
apolipoprotein C-III |
| chrX_+_115567767 | 1.62 |
ENST00000371900.4 |
SLC6A14 |
solute carrier family 6 (amino acid transporter), member 14 |
| chr14_-_25519317 | 1.27 |
ENST00000323944.5 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr2_+_75061108 | 1.24 |
ENST00000290573.2 |
HK2 |
hexokinase 2 |
| chr14_-_25519095 | 1.24 |
ENST00000419632.2 ENST00000358326.2 ENST00000396700.1 ENST00000548724.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr1_-_32801825 | 1.18 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chr16_+_69599899 | 1.08 |
ENST00000567239.1 |
NFAT5 |
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr16_+_69600058 | 1.08 |
ENST00000393742.2 |
NFAT5 |
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr16_+_69599861 | 1.01 |
ENST00000354436.2 |
NFAT5 |
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr2_+_38893047 | 0.86 |
ENST00000272252.5 |
GALM |
galactose mutarotase (aldose 1-epimerase) |
| chr1_+_9599540 | 0.84 |
ENST00000302692.6 |
SLC25A33 |
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr13_-_76056250 | 0.83 |
ENST00000377636.3 ENST00000431480.2 ENST00000377625.2 ENST00000425511.1 |
TBC1D4 |
TBC1 domain family, member 4 |
| chrX_+_152990302 | 0.80 |
ENST00000218104.3 |
ABCD1 |
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr11_-_35440796 | 0.75 |
ENST00000278379.3 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr11_-_35441524 | 0.75 |
ENST00000395750.1 ENST00000449068.1 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr17_+_72428218 | 0.71 |
ENST00000392628.2 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr8_+_17780483 | 0.71 |
ENST00000517730.1 ENST00000518537.1 ENST00000523055.1 ENST00000519253.1 |
PCM1 |
pericentriolar material 1 |
| chr8_+_17780346 | 0.66 |
ENST00000325083.8 |
PCM1 |
pericentriolar material 1 |
| chr2_-_24149977 | 0.66 |
ENST00000238789.5 |
ATAD2B |
ATPase family, AAA domain containing 2B |
| chr2_-_174828892 | 0.64 |
ENST00000418194.2 |
SP3 |
Sp3 transcription factor |
| chr5_-_115152651 | 0.61 |
ENST00000250535.4 |
CDO1 |
cysteine dioxygenase type 1 |
| chr19_+_4969116 | 0.56 |
ENST00000588337.1 ENST00000159111.4 ENST00000381759.4 |
KDM4B |
lysine (K)-specific demethylase 4B |
| chr11_-_47870019 | 0.55 |
ENST00000378460.2 |
NUP160 |
nucleoporin 160kDa |
| chr11_-_47869865 | 0.55 |
ENST00000530326.1 ENST00000532747.1 |
NUP160 |
nucleoporin 160kDa |
| chrX_+_131157290 | 0.52 |
ENST00000394334.2 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chrX_+_131157322 | 0.50 |
ENST00000481105.1 ENST00000354719.6 ENST00000394335.2 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chr1_+_211433275 | 0.47 |
ENST00000367005.4 |
RCOR3 |
REST corepressor 3 |
| chr6_+_37787262 | 0.45 |
ENST00000287218.4 |
ZFAND3 |
zinc finger, AN1-type domain 3 |
| chr17_+_3627185 | 0.45 |
ENST00000325418.4 |
GSG2 |
germ cell associated 2 (haspin) |
| chr3_+_4535025 | 0.44 |
ENST00000302640.8 ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr3_+_171757346 | 0.43 |
ENST00000421757.1 ENST00000415807.2 ENST00000392699.1 |
FNDC3B |
fibronectin type III domain containing 3B |
| chr3_+_52813932 | 0.43 |
ENST00000537050.1 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr6_+_7107999 | 0.42 |
ENST00000491191.1 ENST00000379938.2 ENST00000471433.1 |
RREB1 |
ras responsive element binding protein 1 |
| chr6_+_292051 | 0.41 |
ENST00000344450.5 |
DUSP22 |
dual specificity phosphatase 22 |
| chr6_+_13272904 | 0.40 |
ENST00000379335.3 ENST00000379329.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr5_+_133984462 | 0.39 |
ENST00000398844.2 ENST00000322887.4 |
SEC24A |
SEC24 family member A |
| chr6_+_42531798 | 0.34 |
ENST00000372903.2 ENST00000372899.1 ENST00000372901.1 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
| chr17_-_47755436 | 0.33 |
ENST00000505581.1 ENST00000514121.1 ENST00000393328.2 ENST00000509079.1 ENST00000393331.3 ENST00000347630.2 ENST00000504102.1 |
SPOP |
speckle-type POZ protein |
| chr1_-_114355083 | 0.31 |
ENST00000261441.5 |
RSBN1 |
round spermatid basic protein 1 |
| chr7_-_140624499 | 0.28 |
ENST00000288602.6 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr19_+_36195429 | 0.25 |
ENST00000392197.2 |
ZBTB32 |
zinc finger and BTB domain containing 32 |
| chr1_+_17634689 | 0.24 |
ENST00000375453.1 ENST00000375448.4 |
PADI4 |
peptidyl arginine deiminase, type IV |
| chr17_+_74070853 | 0.24 |
ENST00000329003.3 |
GALR2 |
galanin receptor 2 |
| chr12_-_15942309 | 0.24 |
ENST00000544064.1 ENST00000543523.1 ENST00000536793.1 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
| chr1_+_150122034 | 0.22 |
ENST00000025469.6 ENST00000369124.4 |
PLEKHO1 |
pleckstrin homology domain containing, family O member 1 |
| chr1_-_11866034 | 0.21 |
ENST00000376590.3 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr19_+_50936142 | 0.21 |
ENST00000357701.5 |
MYBPC2 |
myosin binding protein C, fast type |
| chr11_-_118305921 | 0.20 |
ENST00000532619.1 |
RP11-770J1.4 |
RP11-770J1.4 |
| chr2_-_177502254 | 0.19 |
ENST00000339037.3 |
AC017048.3 |
long intergenic non-protein coding RNA 1116 |
| chr17_-_47755338 | 0.18 |
ENST00000508805.1 ENST00000515508.2 ENST00000451526.2 ENST00000507970.1 |
SPOP |
speckle-type POZ protein |
| chr12_+_53773944 | 0.18 |
ENST00000551969.1 ENST00000327443.4 |
SP1 |
Sp1 transcription factor |
| chr20_+_58508817 | 0.17 |
ENST00000358293.3 |
FAM217B |
family with sequence similarity 217, member B |
| chrX_+_128872998 | 0.17 |
ENST00000371106.3 |
XPNPEP2 |
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr1_-_11865982 | 0.16 |
ENST00000418034.1 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr22_+_29601840 | 0.15 |
ENST00000334018.6 ENST00000429226.1 ENST00000404755.3 ENST00000404820.3 ENST00000430127.1 |
EMID1 |
EMI domain containing 1 |
| chr12_-_15942503 | 0.15 |
ENST00000281172.5 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
| chr2_+_232573208 | 0.14 |
ENST00000409115.3 |
PTMA |
prothymosin, alpha |
| chr19_+_36195467 | 0.14 |
ENST00000426659.2 |
ZBTB32 |
zinc finger and BTB domain containing 32 |
| chr9_+_136501478 | 0.14 |
ENST00000393056.2 ENST00000263611.2 |
DBH |
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr3_+_4535155 | 0.14 |
ENST00000544951.1 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr4_+_154387480 | 0.14 |
ENST00000409663.3 ENST00000440693.1 ENST00000409959.3 |
KIAA0922 |
KIAA0922 |
| chr9_+_19230433 | 0.12 |
ENST00000434457.2 ENST00000602925.1 |
DENND4C |
DENN/MADD domain containing 4C |
| chr3_+_155860751 | 0.12 |
ENST00000471742.1 |
KCNAB1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr19_-_11039188 | 0.12 |
ENST00000588347.1 |
YIPF2 |
Yip1 domain family, member 2 |
| chr2_+_232572361 | 0.12 |
ENST00000409321.1 |
PTMA |
prothymosin, alpha |
| chr3_+_49591881 | 0.11 |
ENST00000296452.4 |
BSN |
bassoon presynaptic cytomatrix protein |
| chr4_-_82393009 | 0.11 |
ENST00000436139.2 |
RASGEF1B |
RasGEF domain family, member 1B |
| chr9_-_34662651 | 0.10 |
ENST00000259631.4 |
CCL27 |
chemokine (C-C motif) ligand 27 |
| chrX_+_51636629 | 0.10 |
ENST00000375722.1 ENST00000326587.7 ENST00000375695.2 |
MAGED1 |
melanoma antigen family D, 1 |
| chr2_+_232573222 | 0.08 |
ENST00000341369.7 ENST00000409683.1 |
PTMA |
prothymosin, alpha |
| chr11_+_64009072 | 0.08 |
ENST00000535135.1 ENST00000394540.3 |
FKBP2 |
FK506 binding protein 2, 13kDa |
| chr15_+_27112380 | 0.08 |
ENST00000554596.1 |
GABRA5 |
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr15_+_77287426 | 0.08 |
ENST00000558012.1 ENST00000267939.5 ENST00000379595.3 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
| chr1_-_11865351 | 0.08 |
ENST00000413656.1 ENST00000376585.1 ENST00000423400.1 ENST00000431243.1 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr3_+_105086056 | 0.07 |
ENST00000472644.2 |
ALCAM |
activated leukocyte cell adhesion molecule |
| chr19_-_46389359 | 0.07 |
ENST00000302165.3 |
IRF2BP1 |
interferon regulatory factor 2 binding protein 1 |
| chr6_+_7107830 | 0.07 |
ENST00000379933.3 |
RREB1 |
ras responsive element binding protein 1 |
| chr17_+_29158962 | 0.06 |
ENST00000321990.4 |
ATAD5 |
ATPase family, AAA domain containing 5 |
| chr15_+_27112296 | 0.06 |
ENST00000554038.1 |
GABRA5 |
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr1_-_201346761 | 0.06 |
ENST00000455702.1 ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2 |
troponin T type 2 (cardiac) |
| chr17_+_37824217 | 0.05 |
ENST00000394246.1 |
PNMT |
phenylethanolamine N-methyltransferase |
| chr13_-_27334879 | 0.05 |
ENST00000405846.3 |
GPR12 |
G protein-coupled receptor 12 |
| chr2_+_74741569 | 0.05 |
ENST00000233638.7 |
TLX2 |
T-cell leukemia homeobox 2 |
| chr2_+_239335449 | 0.05 |
ENST00000264607.4 |
ASB1 |
ankyrin repeat and SOCS box containing 1 |
| chr9_+_35605274 | 0.05 |
ENST00000336395.5 |
TESK1 |
testis-specific kinase 1 |
| chr17_+_37824700 | 0.04 |
ENST00000581428.1 |
PNMT |
phenylethanolamine N-methyltransferase |
| chr17_-_31620006 | 0.04 |
ENST00000225823.2 |
ASIC2 |
acid-sensing (proton-gated) ion channel 2 |
| chr14_-_64010046 | 0.04 |
ENST00000337537.3 |
PPP2R5E |
protein phosphatase 2, regulatory subunit B', epsilon isoform |
| chr11_-_65325430 | 0.04 |
ENST00000322147.4 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
| chr11_-_113345995 | 0.03 |
ENST00000355319.2 ENST00000542616.1 |
DRD2 |
dopamine receptor D2 |
| chr3_+_110790590 | 0.03 |
ENST00000485303.1 |
PVRL3 |
poliovirus receptor-related 3 |
| chr16_+_56623433 | 0.02 |
ENST00000570176.1 |
MT3 |
metallothionein 3 |
| chr9_-_21077939 | 0.02 |
ENST00000380232.2 |
IFNB1 |
interferon, beta 1, fibroblast |
| chr1_+_9005917 | 0.01 |
ENST00000549778.1 ENST00000480186.3 ENST00000377443.2 ENST00000377436.3 ENST00000377442.2 |
CA6 |
carbonic anhydrase VI |
| chr2_-_241831424 | 0.00 |
ENST00000402775.2 ENST00000307486.8 |
C2orf54 |
chromosome 2 open reading frame 54 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.8 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.5 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.4 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 2.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.2 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 1.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.4 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 1.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 1.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0060621 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 0.4 | 0.8 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.3 | 1.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.3 | 0.9 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.3 | 0.8 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.3 | 2.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.2 | 0.8 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 1.5 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 1.2 | GO:1901526 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.2 | 0.5 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.2 | 0.6 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.1 | 0.6 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.6 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.4 | GO:0033274 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.5 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.7 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 3.2 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 0.6 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.3 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.1 | 0.2 | GO:1900239 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.1 | 0.3 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.4 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.4 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 1.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0042418 | epinephrine biosynthetic process(GO:0042418) |
| 0.0 | 1.1 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0042421 | norepinephrine biosynthetic process(GO:0042421) |
| 0.0 | 0.4 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.2 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding(GO:0032971) regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.2 | GO:1901018 | positive regulation of potassium ion transmembrane transporter activity(GO:1901018) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.6 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.3 | 0.8 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 1.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 0.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 1.2 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 2.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.8 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.4 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 1.6 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.9 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.2 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.4 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 1.1 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0004603 | phenylethanolamine N-methyltransferase activity(GO:0004603) |
| 0.0 | 0.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |