Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
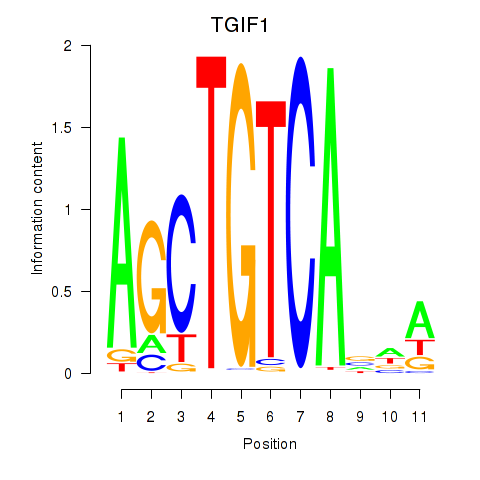
Results for TGIF1
Z-value: 1.40
Transcription factors associated with TGIF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TGIF1
|
ENSG00000177426.16 | TGIF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TGIF1 | hg19_v2_chr18_+_3451646_3451722 | -0.29 | 2.8e-01 | Click! |
Activity profile of TGIF1 motif
Sorted Z-values of TGIF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TGIF1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_33359646 | 3.70 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_33359687 | 3.68 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr3_-_127541679 | 3.45 |
ENST00000265052.5 |
MGLL |
monoglyceride lipase |
| chr4_+_55095264 | 3.45 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr3_-_127541194 | 3.06 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr3_-_127542021 | 2.80 |
ENST00000434178.2 |
MGLL |
monoglyceride lipase |
| chr3_-_127542051 | 2.79 |
ENST00000398104.1 |
MGLL |
monoglyceride lipase |
| chr18_+_54318616 | 2.77 |
ENST00000254442.3 |
WDR7 |
WD repeat domain 7 |
| chr18_+_54318566 | 2.62 |
ENST00000589935.1 ENST00000357574.3 |
WDR7 |
WD repeat domain 7 |
| chr15_-_48937982 | 2.53 |
ENST00000316623.5 |
FBN1 |
fibrillin 1 |
| chr17_+_68165657 | 2.13 |
ENST00000243457.3 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr9_+_125133315 | 2.08 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr3_+_45067659 | 2.07 |
ENST00000296130.4 |
CLEC3B |
C-type lectin domain family 3, member B |
| chr22_-_37640277 | 2.02 |
ENST00000401529.3 ENST00000249071.6 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr18_-_52969844 | 1.93 |
ENST00000561831.3 |
TCF4 |
transcription factor 4 |
| chr17_-_46623441 | 1.86 |
ENST00000330070.4 |
HOXB2 |
homeobox B2 |
| chr19_-_36643329 | 1.84 |
ENST00000589154.1 |
COX7A1 |
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr7_-_93520259 | 1.82 |
ENST00000222543.5 |
TFPI2 |
tissue factor pathway inhibitor 2 |
| chr5_+_141348640 | 1.75 |
ENST00000540015.1 ENST00000506938.1 ENST00000394514.2 ENST00000512565.1 ENST00000394515.3 |
RNF14 |
ring finger protein 14 |
| chr18_-_52989525 | 1.75 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr13_-_38172863 | 1.70 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr4_+_77870960 | 1.62 |
ENST00000505788.1 ENST00000510515.1 ENST00000504637.1 |
SEPT11 |
septin 11 |
| chr9_+_125132803 | 1.61 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr7_-_137028534 | 1.58 |
ENST00000348225.2 |
PTN |
pleiotrophin |
| chrX_-_48931648 | 1.58 |
ENST00000376386.3 ENST00000376390.4 |
PRAF2 |
PRA1 domain family, member 2 |
| chr7_-_137028498 | 1.57 |
ENST00000393083.2 |
PTN |
pleiotrophin |
| chr5_+_141348721 | 1.56 |
ENST00000507163.1 ENST00000394519.1 |
RNF14 |
ring finger protein 14 |
| chr11_+_12132117 | 1.51 |
ENST00000256194.4 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr21_+_30502806 | 1.50 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr5_-_94620239 | 1.48 |
ENST00000515393.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr11_+_6411636 | 1.47 |
ENST00000299397.3 ENST00000356761.2 ENST00000342245.4 |
SMPD1 |
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr7_-_93520191 | 1.44 |
ENST00000545378.1 |
TFPI2 |
tissue factor pathway inhibitor 2 |
| chr14_-_100842588 | 1.42 |
ENST00000556645.1 ENST00000556209.1 ENST00000556504.1 ENST00000556435.1 ENST00000554772.1 ENST00000553581.1 ENST00000553769.2 ENST00000554605.1 ENST00000557722.1 ENST00000553413.1 ENST00000553524.1 ENST00000358655.4 |
WARS |
tryptophanyl-tRNA synthetase |
| chr1_+_163038565 | 1.41 |
ENST00000421743.2 |
RGS4 |
regulator of G-protein signaling 4 |
| chr17_-_76899275 | 1.40 |
ENST00000322630.2 ENST00000586713.1 |
DDC8 |
Protein DDC8 homolog |
| chr9_+_116207007 | 1.38 |
ENST00000374140.2 |
RGS3 |
regulator of G-protein signaling 3 |
| chr5_+_76011868 | 1.32 |
ENST00000319211.4 |
F2R |
coagulation factor II (thrombin) receptor |
| chr2_-_235405168 | 1.31 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr8_-_93029865 | 1.25 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr18_-_21891460 | 1.24 |
ENST00000357041.4 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr9_-_16727978 | 1.22 |
ENST00000418777.1 ENST00000468187.2 |
BNC2 |
basonuclin 2 |
| chr1_+_186798073 | 1.20 |
ENST00000367466.3 ENST00000442353.2 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr18_+_66465302 | 1.18 |
ENST00000360242.5 ENST00000358653.5 |
CCDC102B |
coiled-coil domain containing 102B |
| chr11_-_64052111 | 1.16 |
ENST00000394532.3 ENST00000394531.3 ENST00000309032.3 |
BAD |
BCL2-associated agonist of cell death |
| chr8_+_22446763 | 1.14 |
ENST00000450780.2 ENST00000430850.2 ENST00000447849.1 |
AC037459.4 |
Uncharacterized protein |
| chr16_+_31044413 | 1.14 |
ENST00000394998.1 |
STX4 |
syntaxin 4 |
| chr3_+_112280857 | 1.14 |
ENST00000492406.1 ENST00000468642.1 |
SLC35A5 |
solute carrier family 35, member A5 |
| chr3_+_69788576 | 1.11 |
ENST00000352241.4 ENST00000448226.2 |
MITF |
microphthalmia-associated transcription factor |
| chr10_+_70748487 | 1.07 |
ENST00000361983.4 |
KIAA1279 |
KIAA1279 |
| chr1_-_98386543 | 1.07 |
ENST00000423006.2 ENST00000370192.3 ENST00000306031.5 |
DPYD |
dihydropyrimidine dehydrogenase |
| chr8_+_94929110 | 1.06 |
ENST00000520728.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr2_+_33661382 | 1.05 |
ENST00000402538.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr22_-_37640456 | 1.05 |
ENST00000405484.1 ENST00000441619.1 ENST00000406508.1 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr8_+_94929077 | 1.04 |
ENST00000297598.4 ENST00000520614.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr8_-_49834299 | 1.03 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr1_+_221051699 | 0.98 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr17_-_53809473 | 0.95 |
ENST00000575734.1 |
TMEM100 |
transmembrane protein 100 |
| chr8_-_49833978 | 0.95 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr5_+_141348598 | 0.94 |
ENST00000394520.2 ENST00000347642.3 |
RNF14 |
ring finger protein 14 |
| chr7_+_120591170 | 0.94 |
ENST00000431467.1 |
ING3 |
inhibitor of growth family, member 3 |
| chr1_+_150039369 | 0.93 |
ENST00000369130.3 ENST00000369128.5 |
VPS45 |
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr1_-_149889382 | 0.92 |
ENST00000369145.1 ENST00000369146.3 |
SV2A |
synaptic vesicle glycoprotein 2A |
| chr2_+_185463093 | 0.91 |
ENST00000302277.6 |
ZNF804A |
zinc finger protein 804A |
| chr5_+_140868717 | 0.90 |
ENST00000252087.1 |
PCDHGC5 |
protocadherin gamma subfamily C, 5 |
| chr11_+_6411670 | 0.89 |
ENST00000530395.1 ENST00000527275.1 |
SMPD1 |
sphingomyelin phosphodiesterase 1, acid lysosomal |
| chr1_+_101702417 | 0.88 |
ENST00000305352.6 |
S1PR1 |
sphingosine-1-phosphate receptor 1 |
| chr5_+_125758813 | 0.88 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr16_+_31044812 | 0.88 |
ENST00000313843.3 |
STX4 |
syntaxin 4 |
| chrX_+_155110956 | 0.88 |
ENST00000286448.6 ENST00000262640.6 ENST00000460621.1 |
VAMP7 |
vesicle-associated membrane protein 7 |
| chr7_-_94285472 | 0.87 |
ENST00000437425.2 ENST00000447873.1 ENST00000415788.2 |
SGCE |
sarcoglycan, epsilon |
| chr8_-_42065075 | 0.87 |
ENST00000429089.2 ENST00000519510.1 ENST00000429710.2 ENST00000524009.1 |
PLAT |
plasminogen activator, tissue |
| chr1_+_36621697 | 0.87 |
ENST00000373150.4 ENST00000373151.2 |
MAP7D1 |
MAP7 domain containing 1 |
| chr5_+_34757309 | 0.86 |
ENST00000397449.1 |
RAI14 |
retinoic acid induced 14 |
| chr18_-_54318353 | 0.84 |
ENST00000590954.1 ENST00000540155.1 |
TXNL1 |
thioredoxin-like 1 |
| chr10_-_90751038 | 0.84 |
ENST00000458159.1 ENST00000415557.1 ENST00000458208.1 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr9_-_130635741 | 0.82 |
ENST00000223836.10 |
AK1 |
adenylate kinase 1 |
| chr7_+_30951461 | 0.81 |
ENST00000311813.4 |
AQP1 |
aquaporin 1 (Colton blood group) |
| chr19_-_39826639 | 0.81 |
ENST00000602185.1 ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG |
glia maturation factor, gamma |
| chr3_-_123339343 | 0.81 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chr4_-_149365827 | 0.80 |
ENST00000344721.4 |
NR3C2 |
nuclear receptor subfamily 3, group C, member 2 |
| chrX_-_153775426 | 0.79 |
ENST00000393562.2 |
G6PD |
glucose-6-phosphate dehydrogenase |
| chr10_-_79398250 | 0.78 |
ENST00000286627.5 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr8_+_94929969 | 0.78 |
ENST00000517764.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr15_-_55562582 | 0.78 |
ENST00000396307.2 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr17_-_7297519 | 0.77 |
ENST00000576362.1 ENST00000571078.1 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr19_-_10121144 | 0.77 |
ENST00000264828.3 |
COL5A3 |
collagen, type V, alpha 3 |
| chr3_-_18466026 | 0.76 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr17_-_26220366 | 0.76 |
ENST00000460380.2 ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9 RP1-66C13.4 |
LYR motif containing 9 Uncharacterized protein |
| chr3_-_189840223 | 0.75 |
ENST00000427335.2 |
LEPREL1 |
leprecan-like 1 |
| chr7_-_94285511 | 0.75 |
ENST00000265735.7 |
SGCE |
sarcoglycan, epsilon |
| chr1_-_33168336 | 0.74 |
ENST00000373484.3 |
SYNC |
syncoilin, intermediate filament protein |
| chr9_-_130637244 | 0.74 |
ENST00000373156.1 |
AK1 |
adenylate kinase 1 |
| chr7_-_94285402 | 0.74 |
ENST00000428696.2 ENST00000445866.2 |
SGCE |
sarcoglycan, epsilon |
| chr5_+_32788945 | 0.74 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr11_+_842928 | 0.73 |
ENST00000397408.1 |
TSPAN4 |
tetraspanin 4 |
| chr15_-_55562479 | 0.72 |
ENST00000564609.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr17_-_7297833 | 0.72 |
ENST00000571802.1 ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr9_+_34652164 | 0.72 |
ENST00000441545.2 ENST00000553620.1 |
IL11RA |
interleukin 11 receptor, alpha |
| chr11_-_111781454 | 0.72 |
ENST00000533280.1 |
CRYAB |
crystallin, alpha B |
| chr8_-_82395461 | 0.71 |
ENST00000256104.4 |
FABP4 |
fatty acid binding protein 4, adipocyte |
| chr11_+_842808 | 0.68 |
ENST00000397397.2 ENST00000397411.2 ENST00000397396.1 |
TSPAN4 |
tetraspanin 4 |
| chr5_+_125758865 | 0.68 |
ENST00000542322.1 ENST00000544396.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr9_-_100459639 | 0.67 |
ENST00000375128.4 |
XPA |
xeroderma pigmentosum, complementation group A |
| chr20_-_32308028 | 0.67 |
ENST00000409299.3 ENST00000217398.3 ENST00000344022.3 |
PXMP4 |
peroxisomal membrane protein 4, 24kDa |
| chr11_-_111781610 | 0.67 |
ENST00000525823.1 |
CRYAB |
crystallin, alpha B |
| chr10_-_75255668 | 0.66 |
ENST00000545874.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr17_-_66951474 | 0.66 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr11_-_26743546 | 0.66 |
ENST00000280467.6 ENST00000396005.3 |
SLC5A12 |
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr11_-_842509 | 0.66 |
ENST00000322028.4 |
POLR2L |
polymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa |
| chr1_-_35450897 | 0.64 |
ENST00000373337.3 |
ZMYM6NB |
ZMYM6 neighbor |
| chr8_-_6420930 | 0.63 |
ENST00000325203.5 |
ANGPT2 |
angiopoietin 2 |
| chr7_+_77469439 | 0.63 |
ENST00000450574.1 ENST00000416283.2 ENST00000248550.7 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chr2_-_145275228 | 0.63 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr7_+_134551583 | 0.62 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chr1_-_186649543 | 0.61 |
ENST00000367468.5 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr8_-_6420777 | 0.61 |
ENST00000415216.1 |
ANGPT2 |
angiopoietin 2 |
| chr12_+_104458235 | 0.60 |
ENST00000229330.4 |
HCFC2 |
host cell factor C2 |
| chr10_-_14372870 | 0.60 |
ENST00000357447.2 |
FRMD4A |
FERM domain containing 4A |
| chr8_-_57358432 | 0.60 |
ENST00000517415.1 ENST00000314922.3 |
PENK |
proenkephalin |
| chr6_-_31651817 | 0.60 |
ENST00000375863.3 ENST00000375860.2 |
LY6G5C |
lymphocyte antigen 6 complex, locus G5C |
| chr16_+_50776021 | 0.58 |
ENST00000566679.2 ENST00000564634.1 ENST00000398568.2 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr12_+_62654155 | 0.57 |
ENST00000312635.6 ENST00000393654.3 ENST00000549237.1 |
USP15 |
ubiquitin specific peptidase 15 |
| chr1_+_100111580 | 0.56 |
ENST00000605497.1 |
PALMD |
palmdelphin |
| chr7_-_42276612 | 0.56 |
ENST00000395925.3 ENST00000437480.1 |
GLI3 |
GLI family zinc finger 3 |
| chr14_-_105635090 | 0.55 |
ENST00000331782.3 ENST00000347004.2 |
JAG2 |
jagged 2 |
| chr2_+_242089833 | 0.54 |
ENST00000404405.3 ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7 |
protein phosphatase 1, regulatory subunit 7 |
| chr6_-_134639180 | 0.53 |
ENST00000367858.5 |
SGK1 |
serum/glucocorticoid regulated kinase 1 |
| chr5_-_132112921 | 0.52 |
ENST00000378721.4 ENST00000378701.1 |
SEPT8 |
septin 8 |
| chr3_-_112280709 | 0.52 |
ENST00000402314.2 ENST00000283290.5 ENST00000492886.1 |
ATG3 |
autophagy related 3 |
| chr5_-_132112907 | 0.52 |
ENST00000458488.2 |
SEPT8 |
septin 8 |
| chr1_-_193075180 | 0.52 |
ENST00000367440.3 |
GLRX2 |
glutaredoxin 2 |
| chr22_+_41347363 | 0.51 |
ENST00000216225.8 |
RBX1 |
ring-box 1, E3 ubiquitin protein ligase |
| chr10_+_6244829 | 0.50 |
ENST00000317350.4 ENST00000379785.1 ENST00000379782.3 ENST00000360521.2 ENST00000379775.4 |
PFKFB3 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr1_-_150669500 | 0.50 |
ENST00000271732.3 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr12_-_4647606 | 0.49 |
ENST00000261250.3 ENST00000541014.1 ENST00000545746.1 ENST00000542080.1 |
C12orf4 |
chromosome 12 open reading frame 4 |
| chr14_+_90864504 | 0.49 |
ENST00000544280.2 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
| chr10_-_48438974 | 0.49 |
ENST00000224605.2 |
GDF10 |
growth differentiation factor 10 |
| chr8_+_28174649 | 0.49 |
ENST00000301908.3 |
PNOC |
prepronociceptin |
| chr22_+_39052632 | 0.48 |
ENST00000411557.1 ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1 |
chibby homolog 1 (Drosophila) |
| chr11_+_128634589 | 0.47 |
ENST00000281428.8 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr15_-_37392086 | 0.47 |
ENST00000561208.1 |
MEIS2 |
Meis homeobox 2 |
| chr12_+_59989791 | 0.47 |
ENST00000552432.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr2_-_172750733 | 0.47 |
ENST00000392592.4 ENST00000422440.2 |
SLC25A12 |
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr17_+_40985407 | 0.46 |
ENST00000586114.1 ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3 |
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr12_-_49318715 | 0.45 |
ENST00000444214.2 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
| chr17_-_7145475 | 0.43 |
ENST00000571129.1 ENST00000571253.1 ENST00000573928.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr11_-_111781554 | 0.43 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chr10_+_1102721 | 0.43 |
ENST00000263150.4 |
WDR37 |
WD repeat domain 37 |
| chr11_+_71938925 | 0.41 |
ENST00000538751.1 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
| chr16_-_53537105 | 0.41 |
ENST00000568596.1 ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP |
AKT interacting protein |
| chr11_+_67776012 | 0.40 |
ENST00000539229.1 |
ALDH3B1 |
aldehyde dehydrogenase 3 family, member B1 |
| chr2_+_201936707 | 0.40 |
ENST00000433898.1 ENST00000454214.1 |
NDUFB3 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr10_+_60028818 | 0.39 |
ENST00000333926.5 |
CISD1 |
CDGSH iron sulfur domain 1 |
| chr13_+_30002741 | 0.39 |
ENST00000380808.2 |
MTUS2 |
microtubule associated tumor suppressor candidate 2 |
| chr11_-_73687997 | 0.39 |
ENST00000545212.1 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr12_-_71003568 | 0.39 |
ENST00000547715.1 ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr6_+_32407619 | 0.39 |
ENST00000395388.2 ENST00000374982.5 |
HLA-DRA |
major histocompatibility complex, class II, DR alpha |
| chr12_+_4714145 | 0.38 |
ENST00000545342.1 |
DYRK4 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr3_-_49823941 | 0.38 |
ENST00000321599.4 ENST00000395238.1 ENST00000468463.1 ENST00000460540.1 |
IP6K1 |
inositol hexakisphosphate kinase 1 |
| chr15_+_75335604 | 0.38 |
ENST00000563393.1 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
| chr1_-_19811132 | 0.38 |
ENST00000433834.1 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
| chr8_+_28174496 | 0.37 |
ENST00000518479.1 |
PNOC |
prepronociceptin |
| chr12_+_62654119 | 0.37 |
ENST00000353364.3 ENST00000549523.1 ENST00000280377.5 |
USP15 |
ubiquitin specific peptidase 15 |
| chr17_-_7145106 | 0.37 |
ENST00000577035.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr10_+_70661014 | 0.37 |
ENST00000373585.3 |
DDX50 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr4_+_52709229 | 0.37 |
ENST00000334635.5 ENST00000381441.3 ENST00000381437.4 |
DCUN1D4 |
DCN1, defective in cullin neddylation 1, domain containing 4 |
| chr22_-_37882395 | 0.36 |
ENST00000416983.3 ENST00000424765.2 ENST00000356998.3 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr8_+_71485681 | 0.36 |
ENST00000391684.1 |
AC120194.1 |
AC120194.1 |
| chr3_+_50388126 | 0.36 |
ENST00000425346.1 ENST00000424512.1 ENST00000232508.5 ENST00000418577.1 ENST00000606589.1 |
CYB561D2 XXcos-LUCA11.5 |
cytochrome b561 family, member D2 Uncharacterized protein |
| chr12_-_88974236 | 0.36 |
ENST00000228280.5 ENST00000552044.1 ENST00000357116.4 |
KITLG |
KIT ligand |
| chr10_-_79398127 | 0.35 |
ENST00000372443.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr14_-_65569186 | 0.35 |
ENST00000555932.1 ENST00000358664.4 ENST00000284165.6 ENST00000358402.4 ENST00000246163.2 ENST00000556979.1 ENST00000555667.1 ENST00000557746.1 ENST00000556443.1 |
MAX |
MYC associated factor X |
| chr8_-_6420565 | 0.35 |
ENST00000338312.6 |
ANGPT2 |
angiopoietin 2 |
| chr12_+_59989918 | 0.34 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr12_+_108908962 | 0.34 |
ENST00000552695.1 ENST00000552758.1 ENST00000361549.2 |
FICD |
FIC domain containing |
| chr1_-_150979333 | 0.34 |
ENST00000312210.5 |
FAM63A |
family with sequence similarity 63, member A |
| chr19_+_49375649 | 0.34 |
ENST00000200453.5 |
PPP1R15A |
protein phosphatase 1, regulatory subunit 15A |
| chr3_-_185641681 | 0.34 |
ENST00000259043.7 |
TRA2B |
transformer 2 beta homolog (Drosophila) |
| chr14_-_64194745 | 0.33 |
ENST00000247225.6 |
SGPP1 |
sphingosine-1-phosphate phosphatase 1 |
| chr16_-_30134524 | 0.33 |
ENST00000395202.1 ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3 |
mitogen-activated protein kinase 3 |
| chr14_+_24867992 | 0.33 |
ENST00000382554.3 |
NYNRIN |
NYN domain and retroviral integrase containing |
| chr1_-_150978953 | 0.33 |
ENST00000493834.2 |
FAM63A |
family with sequence similarity 63, member A |
| chrX_-_13835147 | 0.33 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr4_-_175443484 | 0.32 |
ENST00000514584.1 ENST00000542498.1 ENST00000296521.7 ENST00000422112.2 ENST00000504433.1 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr1_-_11118896 | 0.31 |
ENST00000465788.1 |
SRM |
spermidine synthase |
| chr11_-_6426635 | 0.30 |
ENST00000608645.1 ENST00000608394.1 ENST00000529519.1 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chrX_-_153200676 | 0.30 |
ENST00000464845.1 |
NAA10 |
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
| chr11_-_111794446 | 0.30 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr16_-_57219966 | 0.30 |
ENST00000565760.1 ENST00000309137.8 ENST00000570184.1 ENST00000562324.1 |
FAM192A |
family with sequence similarity 192, member A |
| chr9_-_132805430 | 0.29 |
ENST00000446176.2 ENST00000355681.3 ENST00000420781.1 |
FNBP1 |
formin binding protein 1 |
| chr2_-_71454185 | 0.29 |
ENST00000244221.8 |
PAIP2B |
poly(A) binding protein interacting protein 2B |
| chr13_+_96329381 | 0.29 |
ENST00000602402.1 ENST00000376795.6 |
DNAJC3 |
DnaJ (Hsp40) homolog, subfamily C, member 3 |
| chr9_+_34990219 | 0.28 |
ENST00000541010.1 ENST00000454002.2 ENST00000545841.1 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chrX_+_129473916 | 0.28 |
ENST00000545805.1 ENST00000543953.1 ENST00000218197.5 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr2_+_58134756 | 0.28 |
ENST00000435505.2 ENST00000417641.2 |
VRK2 |
vaccinia related kinase 2 |
| chr21_-_35987438 | 0.28 |
ENST00000313806.4 |
RCAN1 |
regulator of calcineurin 1 |
| chr5_-_132113036 | 0.28 |
ENST00000378706.1 |
SEPT8 |
septin 8 |
| chr6_+_73076432 | 0.27 |
ENST00000414192.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr12_+_56325812 | 0.27 |
ENST00000394147.1 ENST00000551156.1 ENST00000553783.1 ENST00000557080.1 ENST00000432422.3 ENST00000556001.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr8_+_26240414 | 0.27 |
ENST00000380629.2 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 3.5 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 2.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 2.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 7.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 5.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.5 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 13.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 2.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 2.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 3.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 3.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 0.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 2.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 3.8 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 1.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.7 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 1.0 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 1.8 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.0 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 12.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 1.2 | 3.5 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 1.1 | 9.8 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 1.0 | 3.1 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.8 | 1.7 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.7 | 2.0 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.7 | 2.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.6 | 1.9 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.5 | 2.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.5 | 1.5 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.5 | 0.5 | GO:0043300 | regulation of myeloid leukocyte mediated immunity(GO:0002886) regulation of leukocyte degranulation(GO:0043300) |
| 0.5 | 1.8 | GO:0050928 | negative regulation of positive chemotaxis(GO:0050928) |
| 0.4 | 1.3 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.4 | 1.1 | GO:0019482 | purine nucleobase catabolic process(GO:0006145) beta-alanine metabolic process(GO:0019482) |
| 0.3 | 2.4 | GO:0023021 | termination of signal transduction(GO:0023021) |
| 0.3 | 1.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.3 | 1.2 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.3 | 2.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.3 | 0.9 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.3 | 1.2 | GO:0052330 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.3 | 1.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 | 3.1 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.3 | 0.8 | GO:0072019 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.3 | 0.8 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.2 | 1.5 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 0.9 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.2 | 0.6 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 1.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.2 | 0.8 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 0.6 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.2 | 0.6 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 2.9 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 0.8 | GO:0072144 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.2 | 1.5 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.2 | 0.6 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 0.6 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.2 | 0.5 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 0.6 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 1.0 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 2.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 1.0 | GO:0051126 | negative regulation of actin nucleation(GO:0051126) |
| 0.1 | 0.4 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.1 | 2.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 1.1 | GO:0045794 | negative regulation of cell volume(GO:0045794) |
| 0.1 | 0.9 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 0.3 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.8 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 3.3 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.4 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.4 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 3.9 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.2 | GO:0002888 | positive regulation of myeloid leukocyte mediated immunity(GO:0002888) positive regulation of leukocyte degranulation(GO:0043302) |
| 0.1 | 0.3 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.3 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.1 | 0.3 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 0.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.3 | GO:0017198 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 2.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.3 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.1 | 0.7 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 1.6 | GO:0015800 | acidic amino acid transport(GO:0015800) L-glutamate transport(GO:0015813) |
| 0.1 | 0.9 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.6 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.5 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.8 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.8 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.7 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0034344 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.3 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.3 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:2001187 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.0 | 0.7 | GO:0050872 | white fat cell differentiation(GO:0050872) cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.9 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.4 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.5 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 1.2 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.4 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 1.1 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 1.6 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.3 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 1.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.2 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0010564 | regulation of cell cycle process(GO:0010564) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.6 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.4 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.2 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 1.3 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.0 | GO:0043400 | cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) |
| 0.0 | 0.1 | GO:1903960 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) plasma membrane long-chain fatty acid transport(GO:0015911) negative regulation of anion transmembrane transport(GO:1903960) |
| 0.0 | 0.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 1.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 5.3 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 3.5 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.9 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.4 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.2 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.2 | GO:0032930 | positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 | 0.7 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.0 | 0.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.9 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.4 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 1.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.5 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 2.4 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.4 | GO:0050436 | microfibril binding(GO:0050436) |
| 1.2 | 3.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 1.1 | 4.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.9 | 12.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.8 | 3.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.5 | 2.0 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.5 | 2.9 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.3 | 2.5 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.3 | 1.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.3 | 3.7 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 0.8 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.3 | 0.8 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.2 | 2.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 1.3 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 1.5 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 0.8 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 1.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 0.9 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 1.8 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 4.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 2.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.5 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.1 | 0.3 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.5 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 0.5 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.7 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 1.1 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.1 | 0.3 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.9 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 2.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.2 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.8 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 1.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 1.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.3 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 1.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 1.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 2.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 1.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.4 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 3.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 2.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 1.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 2.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 2.9 | GO:0019887 | protein kinase regulator activity(GO:0019887) |
| 0.0 | 0.3 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.8 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.4 | 2.1 | GO:0001652 | granular component(GO:0001652) |
| 0.3 | 2.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 0.8 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.3 | 0.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 2.0 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.2 | 0.9 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 0.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 0.5 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.1 | 2.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 1.5 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.3 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 0.6 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 0.7 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.8 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 3.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 7.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.5 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 1.9 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 2.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 3.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 11.0 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 4.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 2.5 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 1.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 2.3 | GO:0044853 | plasma membrane raft(GO:0044853) |
| 0.0 | 2.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.8 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 4.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 1.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.0 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 4.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.8 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.2 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |