Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for THRB
Z-value: 0.62
Transcription factors associated with THRB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
THRB
|
ENSG00000151090.13 | THRB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| THRB | hg19_v2_chr3_-_24207039_24207086, hg19_v2_chr3_-_24536253_24536328 | 0.42 | 1.0e-01 | Click! |
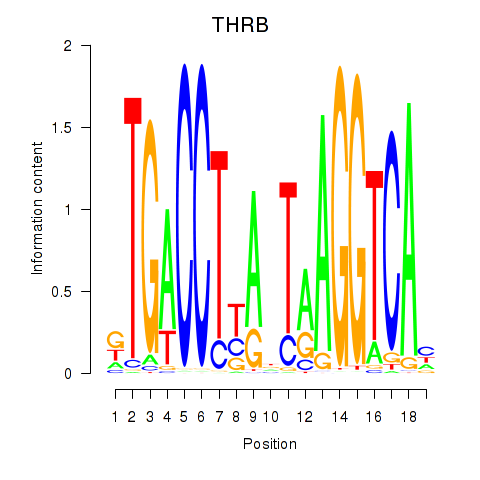
Activity profile of THRB motif
Sorted Z-values of THRB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of THRB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_21810765 | 2.79 |
ENST00000450584.1 ENST00000350669.1 |
LDHB |
lactate dehydrogenase B |
| chr12_-_21810726 | 2.78 |
ENST00000396076.1 |
LDHB |
lactate dehydrogenase B |
| chr20_+_37434329 | 2.29 |
ENST00000299824.1 ENST00000373331.2 |
PPP1R16B |
protein phosphatase 1, regulatory subunit 16B |
| chr8_+_28174649 | 0.90 |
ENST00000301908.3 |
PNOC |
prepronociceptin |
| chr10_+_60145155 | 0.89 |
ENST00000373895.3 |
TFAM |
transcription factor A, mitochondrial |
| chr8_-_124037890 | 0.80 |
ENST00000519018.1 ENST00000523036.1 |
DERL1 |
derlin 1 |
| chr6_-_99842041 | 0.80 |
ENST00000254759.3 ENST00000369242.1 |
COQ3 |
coenzyme Q3 methyltransferase |
| chr10_+_62538089 | 0.79 |
ENST00000519078.2 ENST00000395284.3 ENST00000316629.4 |
CDK1 |
cyclin-dependent kinase 1 |
| chr10_+_62538248 | 0.76 |
ENST00000448257.2 |
CDK1 |
cyclin-dependent kinase 1 |
| chr11_+_73498898 | 0.72 |
ENST00000535529.1 ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48 |
mitochondrial ribosomal protein L48 |
| chr11_+_5617952 | 0.59 |
ENST00000354852.5 |
TRIM6-TRIM34 |
TRIM6-TRIM34 readthrough |
| chr10_+_35456444 | 0.55 |
ENST00000361599.4 |
CREM |
cAMP responsive element modulator |
| chr16_-_69368774 | 0.43 |
ENST00000562949.1 |
RP11-343C2.12 |
Conserved oligomeric Golgi complex subunit 8 |
| chr18_-_47808120 | 0.41 |
ENST00000587605.1 ENST00000591416.1 ENST00000382948.5 ENST00000269471.5 ENST00000269468.5 ENST00000347968.3 ENST00000436910.1 ENST00000349085.2 |
MBD1 |
methyl-CpG binding domain protein 1 |
| chr8_-_100905925 | 0.38 |
ENST00000518171.1 |
COX6C |
cytochrome c oxidase subunit VIc |
| chr6_+_151042224 | 0.36 |
ENST00000358517.2 |
PLEKHG1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr1_-_207224307 | 0.35 |
ENST00000315927.4 |
YOD1 |
YOD1 deubiquitinase |
| chr8_-_100905850 | 0.33 |
ENST00000520271.1 ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C |
cytochrome c oxidase subunit VIc |
| chr18_-_47808050 | 0.31 |
ENST00000590208.1 |
MBD1 |
methyl-CpG binding domain protein 1 |
| chr11_-_47399942 | 0.29 |
ENST00000227163.4 |
SPI1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr4_+_114214125 | 0.28 |
ENST00000509550.1 |
ANK2 |
ankyrin 2, neuronal |
| chrX_-_1511617 | 0.26 |
ENST00000381401.5 |
SLC25A6 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chr8_+_28174496 | 0.25 |
ENST00000518479.1 |
PNOC |
prepronociceptin |
| chr13_-_24007815 | 0.24 |
ENST00000382298.3 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chrY_-_1461617 | 0.22 |
ENSTR0000381401.5 |
SLC25A6 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
| chrX_+_101854096 | 0.22 |
ENST00000246174.2 ENST00000537008.1 ENST00000541409.1 |
ARMCX5 |
armadillo repeat containing, X-linked 5 |
| chr18_-_47807829 | 0.21 |
ENST00000585672.1 ENST00000457839.2 ENST00000353909.3 ENST00000339998.6 ENST00000398493.1 |
MBD1 |
methyl-CpG binding domain protein 1 |
| chr11_-_64014379 | 0.16 |
ENST00000309318.3 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr11_-_47400078 | 0.15 |
ENST00000378538.3 |
SPI1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr8_-_100905363 | 0.15 |
ENST00000524245.1 |
COX6C |
cytochrome c oxidase subunit VIc |
| chr8_+_22102626 | 0.14 |
ENST00000519237.1 ENST00000397802.4 |
POLR3D |
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa |
| chr8_+_22102611 | 0.14 |
ENST00000306433.4 |
POLR3D |
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa |
| chr11_-_45307817 | 0.12 |
ENST00000020926.3 |
SYT13 |
synaptotagmin XIII |
| chr1_-_113498943 | 0.11 |
ENST00000369626.3 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr7_+_102937869 | 0.10 |
ENST00000249269.4 ENST00000428154.1 ENST00000420236.2 |
PMPCB |
peptidase (mitochondrial processing) beta |
| chr11_+_46366918 | 0.07 |
ENST00000528615.1 ENST00000395574.3 |
DGKZ |
diacylglycerol kinase, zeta |
| chr3_+_50606901 | 0.05 |
ENST00000455834.1 |
HEMK1 |
HemK methyltransferase family member 1 |
| chr3_+_50606577 | 0.02 |
ENST00000434410.1 ENST00000232854.4 |
HEMK1 |
HemK methyltransferase family member 1 |
| chr7_-_38394118 | 0.02 |
ENST00000390345.2 |
TRGV4 |
T cell receptor gamma variable 4 |
| chr1_-_154164534 | 0.01 |
ENST00000271850.7 ENST00000368530.2 |
TPM3 |
tropomyosin 3 |
| chr7_-_100253993 | 0.00 |
ENST00000461605.1 ENST00000160382.5 |
ACTL6B |
actin-like 6B |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 0.8 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.0 | 0.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 5.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 1.6 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.4 | 5.6 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.3 | 1.6 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.2 | 0.9 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 0.5 | GO:0052510 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.1 | 0.8 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 | 0.4 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.3 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.9 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.1 | 0.8 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.0 | 0.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.0 | 0.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.6 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 0.9 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.2 | 1.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.5 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.1 | 0.4 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.9 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.1 | 1.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.8 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.8 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.9 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 2.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |