Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
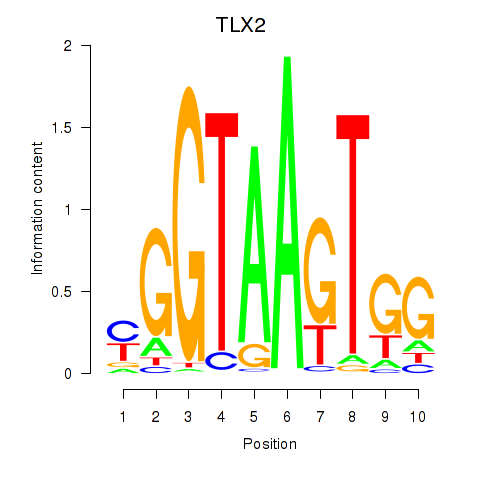
Results for TLX2
Z-value: 1.41
Transcription factors associated with TLX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TLX2
|
ENSG00000115297.9 | TLX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TLX2 | hg19_v2_chr2_+_74741569_74741620 | 0.40 | 1.3e-01 | Click! |
Activity profile of TLX2 motif
Sorted Z-values of TLX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of TLX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_47173994 | 3.44 |
ENST00000414655.2 ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1 LINC00842 |
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr10_+_47746929 | 3.10 |
ENST00000340243.6 ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2 AL603965.1 |
annexin A8-like 2 Protein LOC100996760 |
| chr3_-_182880541 | 2.80 |
ENST00000470251.1 ENST00000265598.3 |
LAMP3 |
lysosomal-associated membrane protein 3 |
| chr10_+_48255253 | 2.56 |
ENST00000357718.4 ENST00000344416.5 ENST00000456111.2 ENST00000374258.3 |
ANXA8 AL591684.1 |
annexin A8 Protein LOC100996760 |
| chr6_+_116692102 | 2.53 |
ENST00000359564.2 |
DSE |
dermatan sulfate epimerase |
| chr11_+_35160709 | 2.42 |
ENST00000415148.2 ENST00000433354.2 ENST00000449691.2 ENST00000437706.2 ENST00000360158.4 ENST00000428726.2 ENST00000526669.2 ENST00000433892.2 ENST00000278386.6 ENST00000434472.2 ENST00000352818.4 ENST00000442151.2 |
CD44 |
CD44 molecule (Indian blood group) |
| chr8_+_86376081 | 2.37 |
ENST00000285379.5 |
CA2 |
carbonic anhydrase II |
| chr6_-_32498046 | 2.17 |
ENST00000374975.3 |
HLA-DRB5 |
major histocompatibility complex, class II, DR beta 5 |
| chr11_+_60223225 | 1.97 |
ENST00000524807.1 ENST00000345732.4 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr1_-_153363452 | 1.97 |
ENST00000368732.1 ENST00000368733.3 |
S100A8 |
S100 calcium binding protein A8 |
| chr16_-_11681023 | 1.93 |
ENST00000570904.1 ENST00000574701.1 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr16_-_11680791 | 1.81 |
ENST00000571976.1 ENST00000413364.2 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr3_+_48507621 | 1.76 |
ENST00000456089.1 |
TREX1 |
three prime repair exonuclease 1 |
| chr11_+_60223312 | 1.74 |
ENST00000532491.1 ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr6_-_138428613 | 1.74 |
ENST00000421351.3 |
PERP |
PERP, TP53 apoptosis effector |
| chr1_+_86889769 | 1.54 |
ENST00000370565.4 |
CLCA2 |
chloride channel accessory 2 |
| chr1_-_25291475 | 1.53 |
ENST00000338888.3 ENST00000399916.1 |
RUNX3 |
runt-related transcription factor 3 |
| chr1_+_152881014 | 1.50 |
ENST00000368764.3 ENST00000392667.2 |
IVL |
involucrin |
| chr3_+_48507210 | 1.50 |
ENST00000433541.1 ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1 |
three prime repair exonuclease 1 |
| chr16_-_11680759 | 1.49 |
ENST00000571459.1 ENST00000570798.1 ENST00000572255.1 ENST00000574763.1 ENST00000574703.1 ENST00000571277.1 ENST00000381810.3 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr3_+_121796697 | 1.46 |
ENST00000482356.1 ENST00000393627.2 |
CD86 |
CD86 molecule |
| chr1_-_68698222 | 1.38 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr6_-_32920794 | 1.33 |
ENST00000395305.3 ENST00000395303.3 ENST00000374843.4 ENST00000429234.1 |
HLA-DMA XXbac-BPG181M17.5 |
major histocompatibility complex, class II, DM alpha Uncharacterized protein |
| chr14_-_67859422 | 1.32 |
ENST00000556532.1 |
PLEK2 |
pleckstrin 2 |
| chr1_+_207627697 | 1.31 |
ENST00000458541.2 |
CR2 |
complement component (3d/Epstein Barr virus) receptor 2 |
| chr20_+_44746939 | 1.30 |
ENST00000372276.3 |
CD40 |
CD40 molecule, TNF receptor superfamily member 5 |
| chr12_-_50677255 | 1.29 |
ENST00000551691.1 ENST00000394943.3 ENST00000341247.4 |
LIMA1 |
LIM domain and actin binding 1 |
| chr1_+_153330322 | 1.27 |
ENST00000368738.3 |
S100A9 |
S100 calcium binding protein A9 |
| chr1_+_207627575 | 1.27 |
ENST00000367058.3 ENST00000367057.3 ENST00000367059.3 |
CR2 |
complement component (3d/Epstein Barr virus) receptor 2 |
| chr20_-_1373606 | 1.26 |
ENST00000381715.1 ENST00000439640.2 ENST00000381719.3 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
| chr20_+_44746885 | 1.26 |
ENST00000372285.3 |
CD40 |
CD40 molecule, TNF receptor superfamily member 5 |
| chr4_-_36246060 | 1.24 |
ENST00000303965.4 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr20_-_14318248 | 1.24 |
ENST00000378053.3 ENST00000341420.4 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
| chr11_+_69924639 | 1.24 |
ENST00000538023.1 ENST00000398543.2 |
ANO1 |
anoctamin 1, calcium activated chloride channel |
| chr3_-_107809816 | 1.21 |
ENST00000361309.5 ENST00000355354.7 |
CD47 |
CD47 molecule |
| chr3_+_121774202 | 1.19 |
ENST00000469710.1 ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86 |
CD86 molecule |
| chr6_-_32634425 | 1.16 |
ENST00000399082.3 ENST00000399079.3 ENST00000374943.4 ENST00000434651.2 |
HLA-DQB1 |
major histocompatibility complex, class II, DQ beta 1 |
| chr12_-_95611149 | 1.15 |
ENST00000549499.1 ENST00000343958.4 ENST00000546711.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr20_-_1373682 | 1.13 |
ENST00000381724.3 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
| chr8_+_26435359 | 1.13 |
ENST00000311151.5 |
DPYSL2 |
dihydropyrimidinase-like 2 |
| chr4_-_105416039 | 1.11 |
ENST00000394767.2 |
CXXC4 |
CXXC finger protein 4 |
| chr15_+_41136586 | 1.11 |
ENST00000431806.1 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr7_+_155090271 | 1.09 |
ENST00000476756.1 |
INSIG1 |
insulin induced gene 1 |
| chr17_-_33760164 | 1.07 |
ENST00000445092.1 ENST00000394562.1 ENST00000447040.2 |
SLFN12 |
schlafen family member 12 |
| chr14_+_61789382 | 1.04 |
ENST00000555082.1 |
PRKCH |
protein kinase C, eta |
| chr17_-_33760269 | 1.02 |
ENST00000452764.3 |
SLFN12 |
schlafen family member 12 |
| chr15_-_38856836 | 1.00 |
ENST00000450598.2 ENST00000559830.1 ENST00000558164.1 ENST00000310803.5 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr7_-_120498357 | 0.99 |
ENST00000415871.1 ENST00000222747.3 ENST00000430985.1 |
TSPAN12 |
tetraspanin 12 |
| chr4_-_90758227 | 0.98 |
ENST00000506691.1 ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr19_+_47104553 | 0.96 |
ENST00000598871.1 ENST00000594523.1 |
CALM3 |
calmodulin 3 (phosphorylase kinase, delta) |
| chr19_-_11688447 | 0.95 |
ENST00000590420.1 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr3_+_69134124 | 0.92 |
ENST00000478935.1 |
ARL6IP5 |
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr4_-_90758118 | 0.91 |
ENST00000420646.2 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr1_+_32739733 | 0.91 |
ENST00000333070.4 |
LCK |
lymphocyte-specific protein tyrosine kinase |
| chr15_-_82555000 | 0.90 |
ENST00000557844.1 ENST00000359445.3 ENST00000268206.7 |
EFTUD1 |
elongation factor Tu GTP binding domain containing 1 |
| chr17_-_33759509 | 0.89 |
ENST00000304905.5 |
SLFN12 |
schlafen family member 12 |
| chr3_+_69134080 | 0.89 |
ENST00000273258.3 |
ARL6IP5 |
ADP-ribosylation-like factor 6 interacting protein 5 |
| chr7_+_90225796 | 0.86 |
ENST00000380050.3 |
CDK14 |
cyclin-dependent kinase 14 |
| chr11_+_128563652 | 0.84 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr17_-_46507537 | 0.84 |
ENST00000336915.6 |
SKAP1 |
src kinase associated phosphoprotein 1 |
| chr19_+_44081344 | 0.83 |
ENST00000599207.1 |
PINLYP |
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr12_-_57522813 | 0.82 |
ENST00000556155.1 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr20_+_43104508 | 0.81 |
ENST00000262605.4 ENST00000372904.3 |
TTPAL |
tocopherol (alpha) transfer protein-like |
| chr14_+_65171315 | 0.79 |
ENST00000394691.1 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr2_-_225811747 | 0.78 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr6_-_31550192 | 0.78 |
ENST00000429299.2 ENST00000446745.2 |
LTB |
lymphotoxin beta (TNF superfamily, member 3) |
| chr15_+_81475047 | 0.77 |
ENST00000559388.1 |
IL16 |
interleukin 16 |
| chrX_+_24483338 | 0.77 |
ENST00000379162.4 ENST00000441463.2 |
PDK3 |
pyruvate dehydrogenase kinase, isozyme 3 |
| chr1_-_32169920 | 0.76 |
ENST00000373672.3 ENST00000373668.3 |
COL16A1 |
collagen, type XVI, alpha 1 |
| chr5_-_151138200 | 0.76 |
ENST00000313115.6 ENST00000522710.1 |
ATOX1 |
antioxidant 1 copper chaperone |
| chr1_-_32169761 | 0.76 |
ENST00000271069.6 |
COL16A1 |
collagen, type XVI, alpha 1 |
| chr16_-_24942273 | 0.75 |
ENST00000571406.1 |
ARHGAP17 |
Rho GTPase activating protein 17 |
| chr2_+_102928009 | 0.74 |
ENST00000404917.2 ENST00000447231.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chr2_+_219433281 | 0.71 |
ENST00000273064.6 ENST00000509807.2 ENST00000542068.1 |
RQCD1 |
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr19_-_11688500 | 0.71 |
ENST00000433365.2 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr1_-_31230650 | 0.69 |
ENST00000294507.3 |
LAPTM5 |
lysosomal protein transmembrane 5 |
| chr14_+_65171099 | 0.68 |
ENST00000247226.7 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr3_+_50192537 | 0.67 |
ENST00000002829.3 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr1_-_68698197 | 0.67 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr3_+_50192499 | 0.66 |
ENST00000413852.1 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr22_-_45636650 | 0.66 |
ENST00000336156.5 |
KIAA0930 |
KIAA0930 |
| chr2_+_219110149 | 0.64 |
ENST00000456575.1 |
ARPC2 |
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr10_-_33623564 | 0.63 |
ENST00000374875.1 ENST00000374822.4 |
NRP1 |
neuropilin 1 |
| chr20_-_1373726 | 0.63 |
ENST00000400137.4 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
| chr16_-_24942411 | 0.62 |
ENST00000571843.1 |
ARHGAP17 |
Rho GTPase activating protein 17 |
| chr17_+_7211280 | 0.61 |
ENST00000419711.2 ENST00000571955.1 ENST00000573714.1 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr3_-_48481518 | 0.61 |
ENST00000412398.2 ENST00000395696.1 |
CCDC51 |
coiled-coil domain containing 51 |
| chr14_+_88471468 | 0.60 |
ENST00000267549.3 |
GPR65 |
G protein-coupled receptor 65 |
| chr22_+_43547520 | 0.60 |
ENST00000337554.3 ENST00000396265.3 ENST00000583777.1 |
TSPO |
translocator protein (18kDa) |
| chr3_+_118892362 | 0.59 |
ENST00000497685.1 ENST00000264234.3 |
UPK1B |
uroplakin 1B |
| chr14_+_23340822 | 0.59 |
ENST00000359591.4 |
LRP10 |
low density lipoprotein receptor-related protein 10 |
| chr16_+_66400533 | 0.59 |
ENST00000341529.3 |
CDH5 |
cadherin 5, type 2 (vascular endothelium) |
| chr8_-_55014336 | 0.58 |
ENST00000343231.6 |
LYPLA1 |
lysophospholipase I |
| chr4_+_78078304 | 0.57 |
ENST00000316355.5 ENST00000354403.5 ENST00000502280.1 |
CCNG2 |
cyclin G2 |
| chr19_-_51512804 | 0.57 |
ENST00000594211.1 ENST00000376832.4 |
KLK9 |
kallikrein-related peptidase 9 |
| chr3_-_105588231 | 0.56 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr1_-_85742773 | 0.55 |
ENST00000370580.1 |
BCL10 |
B-cell CLL/lymphoma 10 |
| chr18_+_29077990 | 0.55 |
ENST00000261590.8 |
DSG2 |
desmoglein 2 |
| chr19_-_45953983 | 0.54 |
ENST00000592083.1 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr4_+_169552748 | 0.54 |
ENST00000504519.1 ENST00000512127.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr1_+_152486950 | 0.53 |
ENST00000368790.3 |
CRCT1 |
cysteine-rich C-terminal 1 |
| chr11_-_89223883 | 0.53 |
ENST00000528341.1 |
NOX4 |
NADPH oxidase 4 |
| chr19_+_1067271 | 0.53 |
ENST00000536472.1 ENST00000590214.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr8_+_74903580 | 0.53 |
ENST00000284818.2 ENST00000518893.1 |
LY96 |
lymphocyte antigen 96 |
| chr5_+_159656437 | 0.53 |
ENST00000402432.3 |
FABP6 |
fatty acid binding protein 6, ileal |
| chr20_-_62710832 | 0.52 |
ENST00000395042.1 |
RGS19 |
regulator of G-protein signaling 19 |
| chr4_-_84205905 | 0.52 |
ENST00000311461.7 ENST00000311469.4 ENST00000439031.2 |
COQ2 |
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
| chr19_+_2389784 | 0.50 |
ENST00000332578.3 |
TMPRSS9 |
transmembrane protease, serine 9 |
| chr6_-_143266297 | 0.50 |
ENST00000367603.2 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
| chr22_+_31477296 | 0.49 |
ENST00000426927.1 ENST00000440425.1 ENST00000358743.1 ENST00000347557.2 ENST00000333137.7 |
SMTN |
smoothelin |
| chr19_-_37019562 | 0.49 |
ENST00000523638.1 |
ZNF260 |
zinc finger protein 260 |
| chr7_+_99971129 | 0.49 |
ENST00000394000.2 ENST00000350573.2 |
PILRA |
paired immunoglobin-like type 2 receptor alpha |
| chr19_+_47104493 | 0.49 |
ENST00000291295.9 ENST00000597743.1 |
CALM3 |
calmodulin 3 (phosphorylase kinase, delta) |
| chr8_+_27183033 | 0.48 |
ENST00000420218.2 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr11_-_33743952 | 0.47 |
ENST00000534312.1 |
CD59 |
CD59 molecule, complement regulatory protein |
| chr12_+_12938541 | 0.47 |
ENST00000356591.4 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr6_-_6007200 | 0.46 |
ENST00000244766.2 |
NRN1 |
neuritin 1 |
| chr19_+_1026298 | 0.46 |
ENST00000263097.4 |
CNN2 |
calponin 2 |
| chr7_+_6414128 | 0.46 |
ENST00000348035.4 ENST00000356142.4 |
RAC1 |
ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
| chr9_+_2621798 | 0.46 |
ENST00000382100.3 |
VLDLR |
very low density lipoprotein receptor |
| chr19_+_1026566 | 0.45 |
ENST00000348419.3 ENST00000565096.2 ENST00000562958.2 ENST00000562075.2 ENST00000607102.1 |
CNN2 |
calponin 2 |
| chr8_+_38758737 | 0.45 |
ENST00000521746.1 ENST00000420274.1 |
PLEKHA2 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr1_-_17304771 | 0.45 |
ENST00000375534.3 |
MFAP2 |
microfibrillar-associated protein 2 |
| chr11_-_22851367 | 0.45 |
ENST00000354193.4 |
SVIP |
small VCP/p97-interacting protein |
| chr10_-_50970322 | 0.44 |
ENST00000374103.4 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr8_-_55014415 | 0.44 |
ENST00000522007.1 ENST00000521898.1 ENST00000518546.1 ENST00000316963.3 |
LYPLA1 |
lysophospholipase I |
| chr9_+_116267536 | 0.44 |
ENST00000374136.1 |
RGS3 |
regulator of G-protein signaling 3 |
| chr10_+_64133934 | 0.43 |
ENST00000395254.3 ENST00000395255.3 ENST00000410046.3 |
ZNF365 |
zinc finger protein 365 |
| chr19_+_36203830 | 0.42 |
ENST00000262630.3 |
ZBTB32 |
zinc finger and BTB domain containing 32 |
| chr19_-_37019136 | 0.42 |
ENST00000592282.1 |
ZNF260 |
zinc finger protein 260 |
| chr7_+_89783689 | 0.41 |
ENST00000297205.2 |
STEAP1 |
six transmembrane epithelial antigen of the prostate 1 |
| chr8_+_27182862 | 0.41 |
ENST00000521164.1 ENST00000346049.5 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr11_-_33744256 | 0.41 |
ENST00000415002.2 ENST00000437761.2 ENST00000445143.2 |
CD59 |
CD59 molecule, complement regulatory protein |
| chr1_+_199996702 | 0.41 |
ENST00000367362.3 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
| chr10_+_71075552 | 0.41 |
ENST00000298649.3 |
HK1 |
hexokinase 1 |
| chr13_-_103426081 | 0.41 |
ENST00000376022.1 ENST00000376021.4 |
TEX30 |
testis expressed 30 |
| chr10_-_50970382 | 0.41 |
ENST00000419399.1 ENST00000432695.1 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr7_-_11871815 | 0.41 |
ENST00000423059.4 |
THSD7A |
thrombospondin, type I, domain containing 7A |
| chr16_-_68269971 | 0.40 |
ENST00000565858.1 |
ESRP2 |
epithelial splicing regulatory protein 2 |
| chr16_+_19183671 | 0.40 |
ENST00000562711.2 |
SYT17 |
synaptotagmin XVII |
| chr14_+_76618242 | 0.39 |
ENST00000557542.1 ENST00000557263.1 ENST00000557207.1 ENST00000312858.5 ENST00000261530.7 |
GPATCH2L |
G patch domain containing 2-like |
| chr11_+_123396528 | 0.39 |
ENST00000322282.7 ENST00000529750.1 |
GRAMD1B |
GRAM domain containing 1B |
| chr13_-_103426112 | 0.39 |
ENST00000376032.4 ENST00000376029.3 |
TEX30 |
testis expressed 30 |
| chr9_+_116225999 | 0.39 |
ENST00000317613.6 |
RGS3 |
regulator of G-protein signaling 3 |
| chr1_+_155829286 | 0.38 |
ENST00000368324.4 |
SYT11 |
synaptotagmin XI |
| chr11_+_67070919 | 0.38 |
ENST00000308127.4 ENST00000308298.7 |
SSH3 |
slingshot protein phosphatase 3 |
| chr12_-_54673871 | 0.37 |
ENST00000209875.4 |
CBX5 |
chromobox homolog 5 |
| chr4_+_2814011 | 0.37 |
ENST00000502260.1 ENST00000435136.2 |
SH3BP2 |
SH3-domain binding protein 2 |
| chr17_+_7452336 | 0.36 |
ENST00000293826.4 |
TNFSF12-TNFSF13 |
TNFSF12-TNFSF13 readthrough |
| chr6_-_16761678 | 0.36 |
ENST00000244769.4 ENST00000436367.1 |
ATXN1 |
ataxin 1 |
| chr15_-_72563585 | 0.36 |
ENST00000287196.9 ENST00000260376.7 |
PARP6 |
poly (ADP-ribose) polymerase family, member 6 |
| chr19_-_6767516 | 0.36 |
ENST00000245908.6 |
SH2D3A |
SH2 domain containing 3A |
| chr20_+_56884752 | 0.36 |
ENST00000244040.3 |
RAB22A |
RAB22A, member RAS oncogene family |
| chr17_+_7452442 | 0.35 |
ENST00000557233.1 |
TNFSF12 |
tumor necrosis factor (ligand) superfamily, member 12 |
| chr1_-_154832316 | 0.35 |
ENST00000361147.4 |
KCNN3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr11_+_67071050 | 0.35 |
ENST00000376757.5 |
SSH3 |
slingshot protein phosphatase 3 |
| chr19_-_59066452 | 0.35 |
ENST00000312547.2 |
CHMP2A |
charged multivesicular body protein 2A |
| chr5_-_102455801 | 0.35 |
ENST00000508629.1 ENST00000399004.2 |
GIN1 |
gypsy retrotransposon integrase 1 |
| chr5_+_131409476 | 0.34 |
ENST00000296871.2 |
CSF2 |
colony stimulating factor 2 (granulocyte-macrophage) |
| chr1_-_6526192 | 0.34 |
ENST00000377782.3 ENST00000351959.5 ENST00000356876.3 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
| chr12_+_9144626 | 0.34 |
ENST00000543895.1 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
| chr19_-_59066327 | 0.34 |
ENST00000596708.1 ENST00000601220.1 ENST00000597848.1 |
CHMP2A |
charged multivesicular body protein 2A |
| chr8_-_27630102 | 0.33 |
ENST00000356537.4 ENST00000522915.1 ENST00000539095.1 |
CCDC25 |
coiled-coil domain containing 25 |
| chrX_+_153665248 | 0.33 |
ENST00000447750.2 |
GDI1 |
GDP dissociation inhibitor 1 |
| chr16_+_58283814 | 0.33 |
ENST00000443128.2 ENST00000219299.4 |
CCDC113 |
coiled-coil domain containing 113 |
| chr7_-_32110451 | 0.31 |
ENST00000396191.1 ENST00000396182.2 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr14_+_54863667 | 0.31 |
ENST00000335183.6 |
CDKN3 |
cyclin-dependent kinase inhibitor 3 |
| chr3_-_36986534 | 0.31 |
ENST00000429976.2 ENST00000301807.6 |
TRANK1 |
tetratricopeptide repeat and ankyrin repeat containing 1 |
| chr1_+_160160283 | 0.31 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr1_+_199996733 | 0.30 |
ENST00000236914.3 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
| chr6_+_13272904 | 0.30 |
ENST00000379335.3 ENST00000379329.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr11_-_133826852 | 0.30 |
ENST00000533871.2 ENST00000321016.8 |
IGSF9B |
immunoglobulin superfamily, member 9B |
| chr1_+_16062820 | 0.30 |
ENST00000294454.5 |
SLC25A34 |
solute carrier family 25, member 34 |
| chr9_+_2622085 | 0.30 |
ENST00000382099.2 |
VLDLR |
very low density lipoprotein receptor |
| chr21_+_43639211 | 0.30 |
ENST00000450121.1 ENST00000398449.3 ENST00000361802.2 |
ABCG1 |
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr14_-_106622419 | 0.29 |
ENST00000390604.2 |
IGHV3-16 |
immunoglobulin heavy variable 3-16 (non-functional) |
| chr19_-_6767431 | 0.29 |
ENST00000437152.3 ENST00000597687.1 |
SH2D3A |
SH2 domain containing 3A |
| chr7_-_150777949 | 0.29 |
ENST00000482571.1 |
FASTK |
Fas-activated serine/threonine kinase |
| chr1_-_226076843 | 0.29 |
ENST00000272134.5 |
LEFTY1 |
left-right determination factor 1 |
| chr5_+_133451254 | 0.28 |
ENST00000517851.1 ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
| chr1_+_9711781 | 0.28 |
ENST00000536656.1 ENST00000377346.4 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr7_-_32111009 | 0.28 |
ENST00000396184.3 ENST00000396189.2 ENST00000321453.7 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr19_-_52133588 | 0.28 |
ENST00000570106.2 |
SIGLEC5 |
sialic acid binding Ig-like lectin 5 |
| chr14_+_54863682 | 0.28 |
ENST00000543789.2 ENST00000442975.2 ENST00000458126.2 ENST00000556102.2 |
CDKN3 |
cyclin-dependent kinase inhibitor 3 |
| chr19_+_16435625 | 0.28 |
ENST00000248071.5 ENST00000592003.1 |
KLF2 |
Kruppel-like factor 2 |
| chr7_-_150777920 | 0.28 |
ENST00000353841.2 ENST00000297532.6 |
FASTK |
Fas-activated serine/threonine kinase |
| chr3_-_122233723 | 0.28 |
ENST00000493510.1 ENST00000344337.6 ENST00000476916.1 ENST00000465882.1 |
KPNA1 |
karyopherin alpha 1 (importin alpha 5) |
| chrX_+_151999511 | 0.27 |
ENST00000370274.3 ENST00000440023.1 ENST00000432467.1 |
NSDHL |
NAD(P) dependent steroid dehydrogenase-like |
| chr1_+_160160346 | 0.27 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr21_-_38445470 | 0.27 |
ENST00000399098.1 |
PIGP |
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr12_+_6602517 | 0.27 |
ENST00000315579.5 ENST00000539714.1 |
NCAPD2 |
non-SMC condensin I complex, subunit D2 |
| chr7_+_99971068 | 0.26 |
ENST00000198536.2 ENST00000453419.1 |
PILRA |
paired immunoglobin-like type 2 receptor alpha |
| chr19_-_42931567 | 0.26 |
ENST00000244289.4 |
LIPE |
lipase, hormone-sensitive |
| chr12_+_113229737 | 0.26 |
ENST00000551052.1 ENST00000415485.3 |
RPH3A |
rabphilin 3A homolog (mouse) |
| chr6_+_106546808 | 0.26 |
ENST00000369089.3 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr2_+_10262857 | 0.26 |
ENST00000304567.5 |
RRM2 |
ribonucleotide reductase M2 |
| chr1_+_948803 | 0.24 |
ENST00000379389.4 |
ISG15 |
ISG15 ubiquitin-like modifier |
| chr15_-_34502278 | 0.24 |
ENST00000559515.1 ENST00000256544.3 ENST00000560108.1 ENST00000559462.1 |
KATNBL1 |
katanin p80 subunit B-like 1 |
| chr12_-_33049690 | 0.24 |
ENST00000070846.6 ENST00000340811.4 |
PKP2 |
plakophilin 2 |
| chr8_+_22423168 | 0.24 |
ENST00000518912.1 ENST00000428103.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.6 | 3.0 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.4 | 2.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.3 | 2.8 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.3 | 2.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.3 | 1.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 0.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 4.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 1.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 0.6 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 1.5 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.1 | 0.6 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.9 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.6 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.1 | 2.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.3 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.1 | 0.6 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 1.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 0.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) sorting endosome(GO:0097443) |
| 0.1 | 0.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.9 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 1.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 1.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.9 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 1.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.2 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 1.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.8 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.0 | 6.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.2 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 2.1 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 2.0 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 2.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.0 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.1 | GO:0031410 | cytoplasmic vesicle(GO:0031410) intracellular vesicle(GO:0097708) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.8 | 2.5 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.5 | 3.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.4 | 1.9 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.4 | 1.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.3 | 0.9 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.3 | 0.9 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 2.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 1.0 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.3 | 0.8 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.2 | 1.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 0.7 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.2 | 3.0 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.2 | 1.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 0.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.2 | 2.5 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.2 | 4.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.2 | 0.9 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.2 | 5.2 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 2.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 2.1 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.2 | 0.9 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 1.8 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 1.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 5.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 3.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.4 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.6 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 4.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.8 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 2.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.6 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 0.7 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 2.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 1.7 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.8 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 3.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.6 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.1 | 0.5 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 1.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.4 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 1.0 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.6 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.8 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.8 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 1.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.4 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.5 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 2.3 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 3.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.8 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 2.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 2.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 3.0 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 1.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 7.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.9 | 2.6 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.8 | 2.4 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.7 | 2.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.5 | 1.5 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.5 | 3.2 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.4 | 1.9 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.4 | 2.6 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.4 | 1.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.3 | 1.3 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.3 | 1.7 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.3 | 2.4 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.3 | 0.8 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.3 | 3.0 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.3 | 2.0 | GO:0036484 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.2 | 1.2 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.2 | 2.5 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.2 | 0.7 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.2 | 0.9 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.2 | 0.9 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.2 | 0.8 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.2 | 0.8 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.2 | 0.2 | GO:1902567 | negative regulation of eosinophil activation(GO:1902567) |
| 0.2 | 1.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.2 | 1.8 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.2 | 1.0 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.2 | 1.0 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.2 | 2.4 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 0.3 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.2 | 0.5 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.1 | 0.9 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.4 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 5.4 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 | 1.2 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 1.0 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.4 | GO:1905154 | negative regulation of membrane invagination(GO:1905154) |
| 0.1 | 1.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.6 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.1 | 0.8 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 0.5 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 2.8 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.3 | GO:0060374 | mast cell differentiation(GO:0060374) |
| 0.1 | 0.3 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.5 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) post-embryonic hemopoiesis(GO:0035166) positive regulation of t-circle formation(GO:1904431) |
| 0.1 | 0.8 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.3 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.1 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.1 | 0.9 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.8 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.1 | 2.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 0.2 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.1 | 0.4 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 0.4 | GO:0051136 | regulation of NK T cell differentiation(GO:0051136) |
| 0.1 | 0.1 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.1 | 1.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 3.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 1.2 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.6 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.4 | GO:0072656 | maintenance of protein location in mitochondrion(GO:0072656) |
| 0.1 | 0.2 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.6 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 1.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.3 | GO:0044336 | embryonic genitalia morphogenesis(GO:0030538) canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 0.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.5 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.6 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 0.3 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.3 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:0072143 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.7 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 3.7 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.8 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 1.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 1.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0097501 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 3.2 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 1.2 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.2 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.2 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.5 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 1.3 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 1.5 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.0 | 0.4 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.4 | GO:0000075 | cell cycle checkpoint(GO:0000075) |
| 0.0 | 0.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:1903061 | regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.0 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 2.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.5 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.7 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.0 | GO:0090298 | late nucleophagy(GO:0044805) negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) single-organism membrane invagination(GO:1902534) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.0 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.0 | GO:0021779 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.0 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.4 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.0 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.1 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 2.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 3.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 2.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 2.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 2.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.6 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.8 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 2.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.2 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |