Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
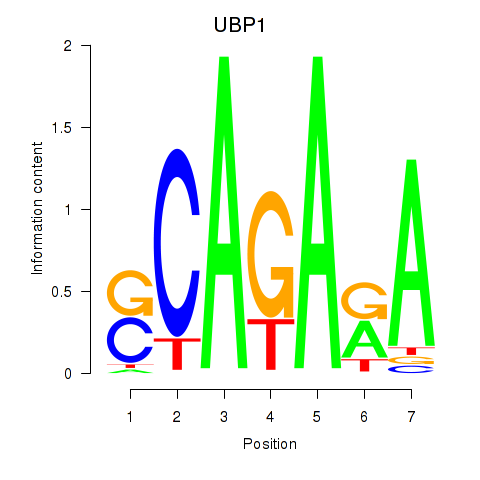
Results for UBP1
Z-value: 0.36
Transcription factors associated with UBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
UBP1
|
ENSG00000153560.7 | UBP1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| UBP1 | hg19_v2_chr3_-_33481835_33481905 | 0.37 | 1.6e-01 | Click! |
Activity profile of UBP1 motif
Sorted Z-values of UBP1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of UBP1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_6588316 | 0.90 |
ENST00000379953.2 |
LY86 |
lymphocyte antigen 86 |
| chr6_+_32709119 | 0.74 |
ENST00000374940.3 |
HLA-DQA2 |
major histocompatibility complex, class II, DQ alpha 2 |
| chr7_+_142498725 | 0.73 |
ENST00000466254.1 |
TRBC2 |
T cell receptor beta constant 2 |
| chr16_-_67970990 | 0.42 |
ENST00000358514.4 |
PSMB10 |
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr11_-_102668879 | 0.37 |
ENST00000315274.6 |
MMP1 |
matrix metallopeptidase 1 (interstitial collagenase) |
| chr1_-_144995074 | 0.29 |
ENST00000534536.1 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr13_-_23949671 | 0.28 |
ENST00000402364.1 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr15_+_81475047 | 0.28 |
ENST00000559388.1 |
IL16 |
interleukin 16 |
| chr11_-_134123142 | 0.28 |
ENST00000392595.2 ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1 |
thymocyte nuclear protein 1 |
| chr12_-_123717643 | 0.28 |
ENST00000541437.1 ENST00000606320.1 |
MPHOSPH9 |
M-phase phosphoprotein 9 |
| chr5_-_111093759 | 0.26 |
ENST00000509979.1 ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP |
neuronal regeneration related protein |
| chr1_-_150208291 | 0.25 |
ENST00000533654.1 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_-_202129105 | 0.25 |
ENST00000367279.4 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
| chr14_+_75988768 | 0.24 |
ENST00000286639.6 |
BATF |
basic leucine zipper transcription factor, ATF-like |
| chr1_-_150208363 | 0.24 |
ENST00000436748.2 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_-_150208412 | 0.23 |
ENST00000532744.1 ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_-_202129704 | 0.23 |
ENST00000476061.1 ENST00000544762.1 ENST00000467283.1 ENST00000464870.1 ENST00000435759.2 ENST00000486116.1 ENST00000543735.1 ENST00000308986.5 ENST00000477625.1 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
| chr1_-_114302086 | 0.23 |
ENST00000369604.1 ENST00000357783.2 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr19_-_51875523 | 0.22 |
ENST00000593572.1 ENST00000595157.1 |
NKG7 |
natural killer cell group 7 sequence |
| chr4_-_70626314 | 0.21 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr8_-_28747424 | 0.21 |
ENST00000523436.1 ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9 |
integrator complex subunit 9 |
| chr1_-_150208498 | 0.20 |
ENST00000314136.8 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr8_-_28747717 | 0.19 |
ENST00000416984.2 |
INTS9 |
integrator complex subunit 9 |
| chr9_-_6015607 | 0.18 |
ENST00000259569.5 |
RANBP6 |
RAN binding protein 6 |
| chr4_-_159080806 | 0.18 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr19_-_59066327 | 0.16 |
ENST00000596708.1 ENST00000601220.1 ENST00000597848.1 |
CHMP2A |
charged multivesicular body protein 2A |
| chr6_+_31555045 | 0.16 |
ENST00000396101.3 ENST00000490742.1 |
LST1 |
leukocyte specific transcript 1 |
| chr22_+_22385332 | 0.15 |
ENST00000390282.2 |
IGLV4-69 |
immunoglobulin lambda variable 4-69 |
| chr22_+_38071615 | 0.15 |
ENST00000215909.5 |
LGALS1 |
lectin, galactoside-binding, soluble, 1 |
| chrX_-_70474910 | 0.15 |
ENST00000373988.1 ENST00000373998.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr2_-_89442621 | 0.14 |
ENST00000492167.1 |
IGKV3-20 |
immunoglobulin kappa variable 3-20 |
| chr9_-_35685452 | 0.14 |
ENST00000607559.1 |
TPM2 |
tropomyosin 2 (beta) |
| chr17_-_62493131 | 0.14 |
ENST00000539111.2 |
POLG2 |
polymerase (DNA directed), gamma 2, accessory subunit |
| chr8_+_77593448 | 0.13 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr1_-_185126037 | 0.12 |
ENST00000367506.5 ENST00000367504.3 |
TRMT1L |
tRNA methyltransferase 1 homolog (S. cerevisiae)-like |
| chr10_-_27443155 | 0.12 |
ENST00000427324.1 ENST00000326799.3 |
YME1L1 |
YME1-like 1 ATPase |
| chr19_-_51538148 | 0.12 |
ENST00000319590.4 ENST00000250351.4 |
KLK12 |
kallikrein-related peptidase 12 |
| chr7_+_142829162 | 0.11 |
ENST00000291009.3 |
PIP |
prolactin-induced protein |
| chr10_-_27443294 | 0.11 |
ENST00000396296.3 ENST00000375972.3 ENST00000376016.3 ENST00000491542.2 |
YME1L1 |
YME1-like 1 ATPase |
| chr1_-_150208320 | 0.11 |
ENST00000534220.1 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr3_-_56502375 | 0.11 |
ENST00000288221.6 |
ERC2 |
ELKS/RAB6-interacting/CAST family member 2 |
| chr1_-_168698433 | 0.10 |
ENST00000367817.3 |
DPT |
dermatopontin |
| chr1_-_144995002 | 0.10 |
ENST00000369356.4 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr1_+_155108294 | 0.10 |
ENST00000303343.8 ENST00000368404.4 ENST00000368401.5 |
SLC50A1 |
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr17_-_5138099 | 0.09 |
ENST00000571800.1 ENST00000574081.1 ENST00000399600.4 ENST00000574297.1 |
SCIMP |
SLP adaptor and CSK interacting membrane protein |
| chr22_+_22730353 | 0.09 |
ENST00000390296.2 |
IGLV5-45 |
immunoglobulin lambda variable 5-45 |
| chr15_-_77712477 | 0.08 |
ENST00000560626.2 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
| chr3_+_29323043 | 0.08 |
ENST00000452462.1 ENST00000456853.1 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
| chr17_+_32683456 | 0.07 |
ENST00000225844.2 |
CCL13 |
chemokine (C-C motif) ligand 13 |
| chr7_+_101460882 | 0.07 |
ENST00000292535.7 ENST00000549414.2 ENST00000550008.2 ENST00000546411.2 ENST00000556210.1 |
CUX1 |
cut-like homeobox 1 |
| chr14_-_92413353 | 0.07 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr14_-_92413727 | 0.06 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr15_-_101835110 | 0.06 |
ENST00000560496.1 |
SNRPA1 |
small nuclear ribonucleoprotein polypeptide A' |
| chr19_-_8070474 | 0.06 |
ENST00000407627.2 ENST00000593807.1 |
ELAVL1 |
ELAV like RNA binding protein 1 |
| chr17_-_42327236 | 0.06 |
ENST00000399246.2 |
AC003102.1 |
AC003102.1 |
| chr4_-_168155169 | 0.05 |
ENST00000534949.1 ENST00000535728.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr11_-_14521379 | 0.04 |
ENST00000249923.3 ENST00000529866.1 ENST00000439561.2 ENST00000534771.1 |
COPB1 |
coatomer protein complex, subunit beta 1 |
| chr1_-_156217875 | 0.04 |
ENST00000292291.5 |
PAQR6 |
progestin and adipoQ receptor family member VI |
| chrX_+_149887090 | 0.04 |
ENST00000538506.1 |
MTMR1 |
myotubularin related protein 1 |
| chr4_-_100815525 | 0.04 |
ENST00000226522.8 ENST00000499666.2 |
LAMTOR3 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr22_+_22676808 | 0.04 |
ENST00000390290.2 |
IGLV1-51 |
immunoglobulin lambda variable 1-51 |
| chr3_+_111260954 | 0.03 |
ENST00000283285.5 |
CD96 |
CD96 molecule |
| chr5_+_122847781 | 0.03 |
ENST00000395412.1 ENST00000395411.1 ENST00000345990.4 |
CSNK1G3 |
casein kinase 1, gamma 3 |
| chr11_-_113345995 | 0.03 |
ENST00000355319.2 ENST00000542616.1 |
DRD2 |
dopamine receptor D2 |
| chr1_+_47901689 | 0.03 |
ENST00000334793.5 |
FOXD2 |
forkhead box D2 |
| chr9_+_108463234 | 0.03 |
ENST00000374688.1 |
TMEM38B |
transmembrane protein 38B |
| chr4_-_57524061 | 0.03 |
ENST00000508121.1 |
HOPX |
HOP homeobox |
| chrX_+_13587712 | 0.02 |
ENST00000361306.1 ENST00000380602.3 |
EGFL6 |
EGF-like-domain, multiple 6 |
| chr11_+_128761220 | 0.02 |
ENST00000529694.1 |
KCNJ5 |
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr19_-_54784937 | 0.01 |
ENST00000434421.1 ENST00000314446.5 ENST00000391749.4 |
LILRB2 |
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 |
| chr1_-_152332480 | 0.01 |
ENST00000388718.5 |
FLG2 |
filaggrin family member 2 |
| chr2_+_113033164 | 0.01 |
ENST00000409871.1 ENST00000343936.4 |
ZC3H6 |
zinc finger CCCH-type containing 6 |
| chr9_+_15422702 | 0.01 |
ENST00000380821.3 ENST00000421710.1 |
SNAPC3 |
small nuclear RNA activating complex, polypeptide 3, 50kDa |
| chr7_+_29874341 | 0.01 |
ENST00000409290.1 ENST00000242140.5 |
WIPF3 |
WAS/WASL interacting protein family, member 3 |
| chr6_-_29395509 | 0.00 |
ENST00000377147.2 |
OR11A1 |
olfactory receptor, family 11, subfamily A, member 1 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 2.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.2 | 3.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 3.5 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 2.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 1.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.3 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 10.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 2.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.4 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.7 | 2.2 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.7 | 2.2 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.4 | 3.5 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 2.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 1.3 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.3 | 1.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.3 | 1.0 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.2 | 0.6 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 5.4 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.2 | 0.5 | GO:0071409 | negative regulation of muscle hyperplasia(GO:0014740) cellular response to cycloheximide(GO:0071409) |
| 0.2 | 0.5 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.2 | 0.8 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.6 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.5 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.9 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 1.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 2.8 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.1 | 0.5 | GO:0048880 | sensory system development(GO:0048880) |
| 0.1 | 0.9 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 0.3 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.1 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.2 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.3 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.7 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.4 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.4 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.6 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.5 | GO:0031293 | Notch receptor processing(GO:0007220) membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 1.9 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.5 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.4 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.0 | 0.1 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.8 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.3 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.0 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.0 | 4.1 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 1.3 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.7 | 3.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.5 | 5.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 2.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.4 | 2.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.3 | 3.8 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 0.6 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.2 | 0.9 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 0.7 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 1.0 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.1 | 0.7 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.5 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 1.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.4 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 2.8 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.4 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 2.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 1.9 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0015403 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.0 | GO:0046921 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.0 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 1.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 3.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 4.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 3.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.9 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 1.0 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 1.0 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0071751 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 3.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.7 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 2.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.2 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.1 | GO:1901656 | glycoside transport(GO:1901656) |
| 0.0 | 1.0 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0002434 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) response to high density lipoprotein particle(GO:0055099) T cell extravasation(GO:0072683) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) |
| 0.0 | 0.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.4 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |