Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for UCACAGU
Z-value: 0.73
miRNA associated with seed UCACAGU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-27a-3p
|
MIMAT0000084 |
|
hsa-miR-27b-3p
|
MIMAT0000419 |
Activity profile of UCACAGU motif
Sorted Z-values of UCACAGU motif
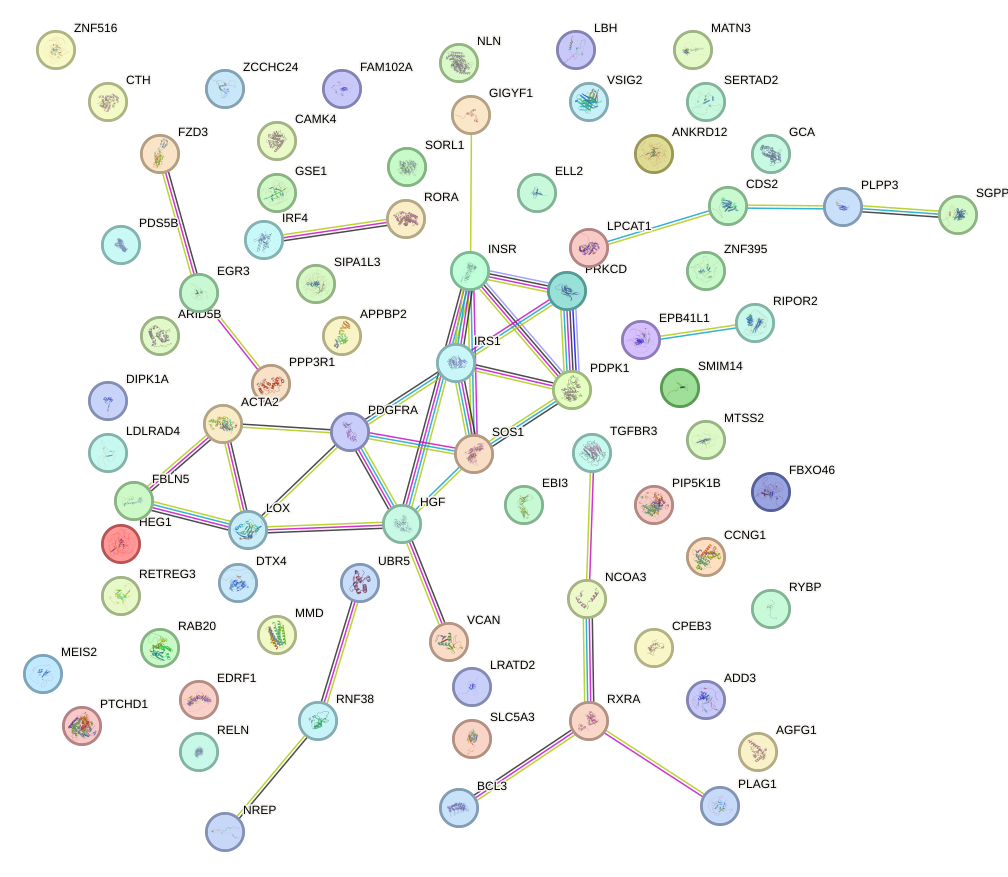
Network of associatons between targets according to the STRING database.
First level regulatory network of UCACAGU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_121322832 | 2.16 |
ENST00000260197.7 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr2_-_38303218 | 1.52 |
ENST00000407341.1 ENST00000260630.3 |
CYP1B1 |
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr7_+_94285637 | 1.17 |
ENST00000482108.1 ENST00000488574.1 |
PEG10 |
paternally expressed 10 |
| chr1_-_92351769 | 1.12 |
ENST00000212355.4 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr1_-_93426998 | 1.07 |
ENST00000370310.4 |
FAM69A |
family with sequence similarity 69, member A |
| chr15_+_81071684 | 0.96 |
ENST00000220244.3 ENST00000394685.3 ENST00000356249.5 |
KIAA1199 |
KIAA1199 |
| chr17_-_53499310 | 0.94 |
ENST00000262065.3 |
MMD |
monocyte to macrophage differentiation-associated |
| chr6_+_391739 | 0.94 |
ENST00000380956.4 |
IRF4 |
interferon regulatory factor 4 |
| chr10_-_90712520 | 0.92 |
ENST00000224784.6 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr1_+_65613217 | 0.88 |
ENST00000545314.1 |
AK4 |
adenylate kinase 4 |
| chr5_-_111093406 | 0.86 |
ENST00000379671.3 |
NREP |
neuronal regeneration related protein |
| chr15_-_61521495 | 0.82 |
ENST00000335670.6 |
RORA |
RAR-related orphan receptor A |
| chr1_-_57045228 | 0.80 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr10_+_63661053 | 0.76 |
ENST00000279873.7 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr19_-_7293942 | 0.74 |
ENST00000341500.5 ENST00000302850.5 |
INSR |
insulin receptor |
| chr9_-_130742792 | 0.74 |
ENST00000373095.1 |
FAM102A |
family with sequence similarity 102, member A |
| chr4_+_55095264 | 0.67 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr7_-_103629963 | 0.67 |
ENST00000428762.1 ENST00000343529.5 ENST00000424685.2 |
RELN |
reelin |
| chr10_+_88516396 | 0.66 |
ENST00000372037.3 |
BMPR1A |
bone morphogenetic protein receptor, type IA |
| chr11_-_115375107 | 0.64 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chr6_+_114178512 | 0.64 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr9_+_137218362 | 0.63 |
ENST00000481739.1 |
RXRA |
retinoid X receptor, alpha |
| chr8_-_28243934 | 0.63 |
ENST00000521185.1 ENST00000520290.1 ENST00000344423.5 |
ZNF395 |
zinc finger protein 395 |
| chr11_+_58939965 | 0.62 |
ENST00000227451.3 |
DTX4 |
deltex homolog 4 (Drosophila) |
| chr1_+_70876891 | 0.61 |
ENST00000411986.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr5_-_1524015 | 0.58 |
ENST00000283415.3 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
| chr3_-_124774802 | 0.55 |
ENST00000311127.4 |
HEG1 |
heart development protein with EGF-like domains 1 |
| chr6_+_138188551 | 0.54 |
ENST00000237289.4 ENST00000433680.1 |
TNFAIP3 |
tumor necrosis factor, alpha-induced protein 3 |
| chr9_+_103790991 | 0.54 |
ENST00000374874.3 |
LPPR1 |
Lipid phosphate phosphatase-related protein type 1 |
| chr18_-_74207146 | 0.53 |
ENST00000443185.2 |
ZNF516 |
zinc finger protein 516 |
| chr1_-_154842741 | 0.52 |
ENST00000271915.4 |
KCNN3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr8_-_22550815 | 0.51 |
ENST00000317216.2 |
EGR3 |
early growth response 3 |
| chr14_-_90085458 | 0.51 |
ENST00000345097.4 ENST00000555855.1 ENST00000555353.1 |
FOXN3 |
forkhead box N3 |
| chr5_+_162864575 | 0.50 |
ENST00000512163.1 ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1 |
cyclin G1 |
| chr1_+_213123915 | 0.49 |
ENST00000366968.4 ENST00000490792.1 |
VASH2 |
vasohibin 2 |
| chr5_-_115910630 | 0.49 |
ENST00000343348.6 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr3_-_72496035 | 0.48 |
ENST00000477973.2 |
RYBP |
RING1 and YY1 binding protein |
| chr12_-_15942309 | 0.48 |
ENST00000544064.1 ENST00000543523.1 ENST00000536793.1 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
| chr2_-_227664474 | 0.46 |
ENST00000305123.5 |
IRS1 |
insulin receptor substrate 1 |
| chr4_-_153457197 | 0.45 |
ENST00000281708.4 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr19_-_46234119 | 0.45 |
ENST00000317683.3 |
FBXO46 |
F-box protein 46 |
| chr7_-_81399438 | 0.42 |
ENST00000222390.5 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr2_+_163200598 | 0.42 |
ENST00000437150.2 ENST00000453113.2 |
GCA |
grancalcin, EF-hand calcium binding protein |
| chr6_-_24911195 | 0.42 |
ENST00000259698.4 |
FAM65B |
family with sequence similarity 65, member B |
| chr2_-_69614373 | 0.41 |
ENST00000361060.5 ENST00000357308.4 |
GFPT1 |
glutamine--fructose-6-phosphate transaminase 1 |
| chr9_+_71320596 | 0.41 |
ENST00000265382.3 |
PIP5K1B |
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr18_+_13218769 | 0.41 |
ENST00000399848.3 ENST00000361205.4 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
| chr5_+_110559784 | 0.40 |
ENST00000282356.4 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
| chr20_+_34700333 | 0.40 |
ENST00000441639.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr16_+_85646763 | 0.39 |
ENST00000411612.1 ENST00000253458.7 |
GSE1 |
Gse1 coiled-coil protein |
| chr5_+_82767284 | 0.38 |
ENST00000265077.3 |
VCAN |
versican |
| chr8_-_57123815 | 0.38 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr13_-_111214015 | 0.37 |
ENST00000267328.3 |
RAB20 |
RAB20, member RAS oncogene family |
| chr14_-_92413727 | 0.37 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr19_+_4229495 | 0.36 |
ENST00000221847.5 |
EBI3 |
Epstein-Barr virus induced 3 |
| chr20_+_46130601 | 0.36 |
ENST00000341724.6 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr9_-_36400213 | 0.36 |
ENST00000259605.6 ENST00000353739.4 |
RNF38 |
ring finger protein 38 |
| chr2_-_20212422 | 0.36 |
ENST00000421259.2 ENST00000407540.3 |
MATN3 |
matrilin 3 |
| chr17_-_58603568 | 0.36 |
ENST00000083182.3 |
APPBP2 |
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr3_+_53195136 | 0.35 |
ENST00000394729.2 ENST00000330452.3 |
PRKCD |
protein kinase C, delta |
| chr5_-_95297678 | 0.35 |
ENST00000237853.4 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr8_-_103425047 | 0.35 |
ENST00000520539.1 |
UBR5 |
ubiquitin protein ligase E3 component n-recognin 5 |
| chr16_+_69599861 | 0.35 |
ENST00000354436.2 |
NFAT5 |
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr10_-_81205373 | 0.34 |
ENST00000372336.3 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
| chr2_-_64881018 | 0.33 |
ENST00000313349.3 |
SERTAD2 |
SERTA domain containing 2 |
| chr2_+_228336849 | 0.33 |
ENST00000409979.2 ENST00000310078.8 |
AGFG1 |
ArfGAP with FG repeats 1 |
| chr14_-_64194745 | 0.33 |
ENST00000247225.6 |
SGPP1 |
sphingosine-1-phosphate phosphatase 1 |
| chr4_-_39640700 | 0.33 |
ENST00000295958.5 |
SMIM14 |
small integral membrane protein 14 |
| chr19_+_45251804 | 0.32 |
ENST00000164227.5 |
BCL3 |
B-cell CLL/lymphoma 3 |
| chr13_+_33160553 | 0.32 |
ENST00000315596.10 |
PDS5B |
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr18_+_55102917 | 0.32 |
ENST00000491143.2 |
ONECUT2 |
one cut homeobox 2 |
| chr8_-_127570603 | 0.32 |
ENST00000304916.3 |
FAM84B |
family with sequence similarity 84, member B |
| chr12_+_60083118 | 0.31 |
ENST00000261187.4 ENST00000543448.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_-_200992827 | 0.31 |
ENST00000332129.2 ENST00000422435.2 |
KIF21B |
kinesin family member 21B |
| chr16_-_70719925 | 0.31 |
ENST00000338779.6 |
MTSS1L |
metastasis suppressor 1-like |
| chr17_-_40761375 | 0.31 |
ENST00000543197.1 ENST00000309428.5 |
FAM134C |
family with sequence similarity 134, member C |
| chr8_+_38758737 | 0.31 |
ENST00000521746.1 ENST00000420274.1 |
PLEKHA2 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chrX_+_23352133 | 0.31 |
ENST00000379361.4 |
PTCHD1 |
patched domain containing 1 |
| chr15_+_73344791 | 0.30 |
ENST00000261908.6 |
NEO1 |
neogenin 1 |
| chr12_-_102874416 | 0.30 |
ENST00000392904.1 ENST00000337514.6 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr21_+_35445827 | 0.30 |
ENST00000608209.1 ENST00000381151.3 |
SLC5A3 SLC5A3 |
sodium/myo-inositol cotransporter solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
| chr2_+_30454390 | 0.30 |
ENST00000395323.3 ENST00000406087.1 ENST00000404397.1 |
LBH |
limb bud and heart development |
| chr10_+_111767720 | 0.29 |
ENST00000356080.4 ENST00000277900.8 |
ADD3 |
adducin 3 (gamma) |
| chrX_-_110655391 | 0.29 |
ENST00000356915.2 ENST00000356220.3 |
DCX |
doublecortin |
| chr16_+_2587998 | 0.29 |
ENST00000441549.3 ENST00000268673.7 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr2_-_68479614 | 0.29 |
ENST00000234310.3 |
PPP3R1 |
protein phosphatase 3, regulatory subunit B, alpha |
| chr16_+_23847339 | 0.28 |
ENST00000303531.7 |
PRKCB |
protein kinase C, beta |
| chr15_-_37390482 | 0.28 |
ENST00000559085.1 ENST00000397624.3 |
MEIS2 |
Meis homeobox 2 |
| chr5_+_65018017 | 0.27 |
ENST00000380985.5 ENST00000502464.1 |
NLN |
neurolysin (metallopeptidase M3 family) |
| chr10_+_127408263 | 0.27 |
ENST00000337623.3 |
C10orf137 |
erythroid differentiation regulatory factor 1 |
| chr8_+_28351707 | 0.27 |
ENST00000537916.1 ENST00000523546.1 ENST00000240093.3 |
FZD3 |
frizzled family receptor 3 |
| chr20_+_5107420 | 0.27 |
ENST00000460006.1 |
CDS2 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr18_+_9136758 | 0.27 |
ENST00000383440.2 ENST00000262126.4 ENST00000577992.1 |
ANKRD12 |
ankyrin repeat domain 12 |
| chr19_+_38397839 | 0.27 |
ENST00000222345.6 |
SIPA1L3 |
signal-induced proliferation-associated 1 like 3 |
| chr5_-_121413974 | 0.27 |
ENST00000231004.4 |
LOX |
lysyl oxidase |
| chr10_-_94003003 | 0.26 |
ENST00000412050.4 |
CPEB3 |
cytoplasmic polyadenylation element binding protein 3 |
| chr3_+_23986748 | 0.26 |
ENST00000312521.4 |
NR1D2 |
nuclear receptor subfamily 1, group D, member 2 |
| chr10_+_104474207 | 0.26 |
ENST00000602831.1 ENST00000369893.5 |
SFXN2 |
sideroflexin 2 |
| chr16_-_11680791 | 0.25 |
ENST00000571976.1 ENST00000413364.2 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr7_+_90225796 | 0.25 |
ENST00000380050.3 |
CDK14 |
cyclin-dependent kinase 14 |
| chr16_-_71758602 | 0.25 |
ENST00000568954.1 |
PHLPP2 |
PH domain and leucine rich repeat protein phosphatase 2 |
| chr3_-_178790057 | 0.25 |
ENST00000311417.2 |
ZMAT3 |
zinc finger, matrin-type 3 |
| chr10_+_112257596 | 0.25 |
ENST00000369583.3 |
DUSP5 |
dual specificity phosphatase 5 |
| chr1_-_149889382 | 0.25 |
ENST00000369145.1 ENST00000369146.3 |
SV2A |
synaptic vesicle glycoprotein 2A |
| chr19_-_49576198 | 0.25 |
ENST00000221444.1 |
KCNA7 |
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr3_+_58223228 | 0.25 |
ENST00000478253.1 ENST00000295962.4 |
ABHD6 |
abhydrolase domain containing 6 |
| chr2_+_169312350 | 0.24 |
ENST00000305747.6 |
CERS6 |
ceramide synthase 6 |
| chr13_-_77900814 | 0.24 |
ENST00000544440.2 |
MYCBP2 |
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr1_-_19229248 | 0.24 |
ENST00000375341.3 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
| chr8_-_37756972 | 0.24 |
ENST00000330843.4 ENST00000522727.1 ENST00000287263.4 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
| chr8_+_26240414 | 0.24 |
ENST00000380629.2 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr8_-_8751068 | 0.24 |
ENST00000276282.6 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
| chr17_-_73389737 | 0.24 |
ENST00000392563.1 |
GRB2 |
growth factor receptor-bound protein 2 |
| chr1_+_192544857 | 0.24 |
ENST00000367459.3 ENST00000469578.2 |
RGS1 |
regulator of G-protein signaling 1 |
| chr3_-_114790179 | 0.23 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr16_+_11762270 | 0.23 |
ENST00000329565.5 |
SNN |
stannin |
| chr5_-_93447333 | 0.23 |
ENST00000395965.3 ENST00000505869.1 ENST00000509163.1 |
FAM172A |
family with sequence similarity 172, member A |
| chr5_-_81046922 | 0.23 |
ENST00000514493.1 ENST00000320672.4 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr10_+_60272814 | 0.23 |
ENST00000373886.3 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
| chr3_+_186330712 | 0.23 |
ENST00000411641.2 ENST00000273784.5 |
AHSG |
alpha-2-HS-glycoprotein |
| chr17_+_30813576 | 0.23 |
ENST00000313401.3 |
CDK5R1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr16_-_73082274 | 0.23 |
ENST00000268489.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr11_-_73309228 | 0.23 |
ENST00000356467.4 ENST00000064778.4 |
FAM168A |
family with sequence similarity 168, member A |
| chr7_-_139876812 | 0.23 |
ENST00000397560.2 |
JHDM1D |
lysine (K)-specific demethylase 7A |
| chr10_-_124768300 | 0.22 |
ENST00000368886.5 |
IKZF5 |
IKAROS family zinc finger 5 (Pegasus) |
| chr12_-_81331697 | 0.22 |
ENST00000552864.1 |
LIN7A |
lin-7 homolog A (C. elegans) |
| chr8_-_41166953 | 0.22 |
ENST00000220772.3 |
SFRP1 |
secreted frizzled-related protein 1 |
| chr3_+_16926441 | 0.22 |
ENST00000418129.2 ENST00000396755.2 |
PLCL2 |
phospholipase C-like 2 |
| chr9_-_127905736 | 0.22 |
ENST00000336505.6 ENST00000373549.4 |
SCAI |
suppressor of cancer cell invasion |
| chr10_+_52751010 | 0.22 |
ENST00000373985.1 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr4_+_113970772 | 0.22 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr11_-_30038490 | 0.21 |
ENST00000328224.6 |
KCNA4 |
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr10_-_15762124 | 0.21 |
ENST00000378076.3 |
ITGA8 |
integrin, alpha 8 |
| chrX_-_46618490 | 0.21 |
ENST00000328306.4 |
SLC9A7 |
solute carrier family 9, subfamily A (NHE7, cation proton antiporter 7), member 7 |
| chr2_+_85198216 | 0.20 |
ENST00000456682.1 ENST00000409785.4 |
KCMF1 |
potassium channel modulatory factor 1 |
| chr6_-_46293378 | 0.20 |
ENST00000330430.6 |
RCAN2 |
regulator of calcineurin 2 |
| chr8_-_94753229 | 0.20 |
ENST00000518597.1 ENST00000399300.2 ENST00000517700.1 |
RBM12B |
RNA binding motif protein 12B |
| chr18_+_2655692 | 0.20 |
ENST00000320876.6 |
SMCHD1 |
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr3_+_107241783 | 0.20 |
ENST00000415149.2 ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX |
bobby sox homolog (Drosophila) |
| chr6_+_126112001 | 0.20 |
ENST00000392477.2 |
NCOA7 |
nuclear receptor coactivator 7 |
| chr1_+_203274639 | 0.20 |
ENST00000290551.4 |
BTG2 |
BTG family, member 2 |
| chr8_-_122653630 | 0.19 |
ENST00000303924.4 |
HAS2 |
hyaluronan synthase 2 |
| chr8_-_98290087 | 0.19 |
ENST00000322128.3 |
TSPYL5 |
TSPY-like 5 |
| chr12_-_106641728 | 0.19 |
ENST00000378026.4 |
CKAP4 |
cytoskeleton-associated protein 4 |
| chr3_-_21792838 | 0.19 |
ENST00000281523.2 |
ZNF385D |
zinc finger protein 385D |
| chr1_-_108507631 | 0.19 |
ENST00000527011.1 ENST00000370056.4 |
VAV3 |
vav 3 guanine nucleotide exchange factor |
| chr2_+_7057523 | 0.19 |
ENST00000320892.6 |
RNF144A |
ring finger protein 144A |
| chr19_+_4969116 | 0.18 |
ENST00000588337.1 ENST00000159111.4 ENST00000381759.4 |
KDM4B |
lysine (K)-specific demethylase 4B |
| chr12_+_862089 | 0.18 |
ENST00000315939.6 ENST00000537687.1 ENST00000447667.2 |
WNK1 |
WNK lysine deficient protein kinase 1 |
| chr2_-_166930131 | 0.18 |
ENST00000303395.4 ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A |
sodium channel, voltage-gated, type I, alpha subunit |
| chr2_-_172290482 | 0.18 |
ENST00000442541.1 ENST00000392599.2 ENST00000375258.4 |
METTL8 |
methyltransferase like 8 |
| chr2_-_39348137 | 0.18 |
ENST00000426016.1 |
SOS1 |
son of sevenless homolog 1 (Drosophila) |
| chr15_+_33010175 | 0.18 |
ENST00000300177.4 ENST00000560677.1 ENST00000560830.1 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chr18_-_31803435 | 0.17 |
ENST00000589544.1 ENST00000269185.4 ENST00000261592.5 |
NOL4 |
nucleolar protein 4 |
| chr22_-_39239987 | 0.17 |
ENST00000333039.2 |
NPTXR |
neuronal pentraxin receptor |
| chr1_+_87380299 | 0.17 |
ENST00000370551.4 ENST00000370550.5 |
HS2ST1 |
heparan sulfate 2-O-sulfotransferase 1 |
| chr16_-_48644061 | 0.16 |
ENST00000262384.3 |
N4BP1 |
NEDD4 binding protein 1 |
| chr11_-_77532050 | 0.16 |
ENST00000308488.6 |
RSF1 |
remodeling and spacing factor 1 |
| chr2_-_24149977 | 0.16 |
ENST00000238789.5 |
ATAD2B |
ATPase family, AAA domain containing 2B |
| chr17_+_46184911 | 0.15 |
ENST00000580219.1 ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11 |
sorting nexin 11 |
| chr1_+_211433275 | 0.15 |
ENST00000367005.4 |
RCOR3 |
REST corepressor 3 |
| chr17_-_76713100 | 0.15 |
ENST00000585509.1 |
CYTH1 |
cytohesin 1 |
| chr3_-_129407535 | 0.15 |
ENST00000432054.2 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
| chr11_-_60674037 | 0.15 |
ENST00000541371.1 ENST00000227524.4 |
PRPF19 |
pre-mRNA processing factor 19 |
| chr12_-_75905374 | 0.15 |
ENST00000438169.2 ENST00000229214.4 |
KRR1 |
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr2_-_232645977 | 0.15 |
ENST00000409772.1 |
PDE6D |
phosphodiesterase 6D, cGMP-specific, rod, delta |
| chr6_+_30524663 | 0.15 |
ENST00000376560.3 |
PRR3 |
proline rich 3 |
| chr19_+_19496624 | 0.15 |
ENST00000494516.2 ENST00000360315.3 ENST00000252577.5 |
GATAD2A |
GATA zinc finger domain containing 2A |
| chr1_-_67896095 | 0.15 |
ENST00000370994.4 |
SERBP1 |
SERPINE1 mRNA binding protein 1 |
| chr3_+_38495333 | 0.15 |
ENST00000352511.4 |
ACVR2B |
activin A receptor, type IIB |
| chr6_+_37787262 | 0.15 |
ENST00000287218.4 |
ZFAND3 |
zinc finger, AN1-type domain 3 |
| chr5_+_133984462 | 0.15 |
ENST00000398844.2 ENST00000322887.4 |
SEC24A |
SEC24 family member A |
| chr3_+_54156664 | 0.15 |
ENST00000474759.1 ENST00000288197.5 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chrX_+_108780062 | 0.15 |
ENST00000372106.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr1_+_39456895 | 0.15 |
ENST00000432648.3 ENST00000446189.2 ENST00000372984.4 |
AKIRIN1 |
akirin 1 |
| chr2_-_165697920 | 0.14 |
ENST00000342193.4 ENST00000375458.2 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr20_-_33999766 | 0.14 |
ENST00000349714.5 ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1 |
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chrX_+_30671476 | 0.14 |
ENST00000378946.3 ENST00000378943.3 ENST00000378945.3 ENST00000427190.1 ENST00000378941.3 |
GK |
glycerol kinase |
| chr10_-_52645416 | 0.14 |
ENST00000374001.2 ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF |
APOBEC1 complementation factor |
| chr11_-_74109422 | 0.14 |
ENST00000298198.4 |
PGM2L1 |
phosphoglucomutase 2-like 1 |
| chr5_+_65440032 | 0.14 |
ENST00000334121.6 |
SREK1 |
splicing regulatory glutamine/lysine-rich protein 1 |
| chr5_+_32585605 | 0.14 |
ENST00000265073.4 ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
| chr17_-_35969409 | 0.14 |
ENST00000394378.2 ENST00000585472.1 ENST00000591288.1 ENST00000502449.2 ENST00000345615.4 ENST00000346661.4 ENST00000585689.1 ENST00000339208.6 |
SYNRG |
synergin, gamma |
| chr2_-_100939195 | 0.14 |
ENST00000393437.3 |
LONRF2 |
LON peptidase N-terminal domain and ring finger 2 |
| chr14_+_71108460 | 0.14 |
ENST00000256367.2 |
TTC9 |
tetratricopeptide repeat domain 9 |
| chr6_+_34759837 | 0.14 |
ENST00000192788.5 ENST00000452449.2 |
UHRF1BP1 |
UHRF1 binding protein 1 |
| chr11_-_67888671 | 0.13 |
ENST00000265689.4 |
CHKA |
choline kinase alpha |
| chr1_+_205473720 | 0.13 |
ENST00000429964.2 ENST00000506784.1 ENST00000360066.2 |
CDK18 |
cyclin-dependent kinase 18 |
| chr18_-_45456930 | 0.13 |
ENST00000262160.6 ENST00000587269.1 |
SMAD2 |
SMAD family member 2 |
| chr12_-_12419703 | 0.13 |
ENST00000543091.1 ENST00000261349.4 |
LRP6 |
low density lipoprotein receptor-related protein 6 |
| chr7_+_74072011 | 0.13 |
ENST00000324896.4 ENST00000353920.4 ENST00000346152.4 ENST00000416070.1 |
GTF2I |
general transcription factor IIi |
| chr13_-_76056250 | 0.13 |
ENST00000377636.3 ENST00000431480.2 ENST00000377625.2 ENST00000425511.1 |
TBC1D4 |
TBC1 domain family, member 4 |
| chr3_-_125313934 | 0.13 |
ENST00000296220.5 |
OSBPL11 |
oxysterol binding protein-like 11 |
| chr16_-_22385901 | 0.13 |
ENST00000268383.2 |
CDR2 |
cerebellar degeneration-related protein 2, 62kDa |
| chr14_-_100070363 | 0.13 |
ENST00000380243.4 |
CCDC85C |
coiled-coil domain containing 85C |
| chr5_-_132073210 | 0.13 |
ENST00000378735.1 ENST00000378746.4 |
KIF3A |
kinesin family member 3A |
| chr9_+_114659046 | 0.13 |
ENST00000374279.3 |
UGCG |
UDP-glucose ceramide glucosyltransferase |
| chr5_-_132948216 | 0.12 |
ENST00000265342.7 |
FSTL4 |
follistatin-like 4 |
| chr13_+_26828275 | 0.12 |
ENST00000381527.3 |
CDK8 |
cyclin-dependent kinase 8 |
| chr3_-_171178157 | 0.12 |
ENST00000465393.1 ENST00000436636.2 ENST00000369326.5 ENST00000538048.1 ENST00000341852.6 |
TNIK |
TRAF2 and NCK interacting kinase |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 1.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 0.9 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 2.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.1 | GO:0070160 | bicellular tight junction(GO:0005923) apical junction complex(GO:0043296) occluding junction(GO:0070160) |
| 0.1 | 0.1 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.1 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.3 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.2 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.1 | 0.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.5 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.7 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.7 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.3 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.3 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.0 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.8 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 0.7 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.2 | 0.9 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 0.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.2 | 0.6 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 0.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.4 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.1 | 2.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 0.8 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.6 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.1 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.3 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 0.3 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 0.7 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 1.5 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.3 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 1.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.0 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 0.3 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.0 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.4 | 1.3 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.4 | 1.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.3 | 1.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.2 | 0.7 | GO:0090291 | negative regulation of osteoclast proliferation(GO:0090291) |
| 0.2 | 0.7 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.2 | 0.7 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.2 | 0.6 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 | 0.6 | GO:2001245 | negative regulation of phospholipid biosynthetic process(GO:0071072) regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.2 | 0.7 | GO:1990535 | transformation of host cell by virus(GO:0019087) neuron projection maintenance(GO:1990535) |
| 0.2 | 0.9 | GO:0072144 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.2 | 0.7 | GO:0097477 | NMDA glutamate receptor clustering(GO:0097114) spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.2 | 0.6 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 0.6 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.9 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 0.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.4 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 0.3 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 0.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.3 | GO:1902809 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.3 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.1 | 0.5 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.4 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.3 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.1 | GO:1902954 | regulation of early endosome to recycling endosome transport(GO:1902954) |
| 0.1 | 0.5 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) |
| 0.1 | 0.4 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.3 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 | 0.6 | GO:0090269 | fibroblast growth factor production(GO:0090269) regulation of fibroblast growth factor production(GO:0090270) |
| 0.1 | 0.2 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.2 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 0.2 | GO:2000729 | positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.2 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 0.3 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.3 | GO:2000035 | positive regulation of somatic stem cell population maintenance(GO:1904674) regulation of stem cell division(GO:2000035) |
| 0.1 | 0.8 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.1 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.1 | 0.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 0.4 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.0 | 0.3 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.2 | GO:1900127 | renal water absorption(GO:0070295) positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.3 | GO:0015791 | polyol transport(GO:0015791) |
| 0.0 | 0.5 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.4 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.2 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.4 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.3 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.0 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.0 | 0.2 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.1 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.3 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.4 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.1 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 1.1 | GO:0016577 | histone demethylation(GO:0016577) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.2 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.1 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.4 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.0 | 0.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.5 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.1 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.3 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.4 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.0 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.0 | 0.0 | GO:2001038 | regulation of cellular response to drug(GO:2001038) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:1901160 | primary amino compound metabolic process(GO:1901160) |
| 0.0 | 0.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.0 | GO:0007497 | posterior midgut development(GO:0007497) |
| 0.0 | 0.0 | GO:0042636 | negative regulation of hair cycle(GO:0042636) |
| 0.0 | 1.1 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 0.1 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.3 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0032912 | negative regulation of transforming growth factor beta2 production(GO:0032912) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.0 | 0.1 | GO:0044254 | angiotensin catabolic process in blood(GO:0002005) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) |
| 0.0 | 0.3 | GO:2000505 | regulation of energy homeostasis(GO:2000505) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.0 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.0 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.1 | GO:0034378 | chylomicron assembly(GO:0034378) |
| 0.0 | 0.1 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.0 | GO:0032909 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.0 | 0.1 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.1 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.6 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.8 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.7 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.1 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.5 | PID PI3KCI PATHWAY | Class I PI3K signaling events |