Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
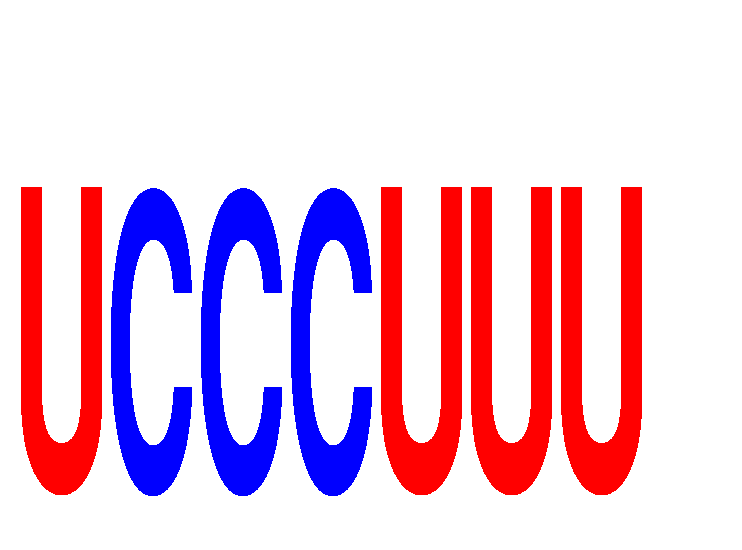
Results for UCCCUUU
Z-value: 0.39
miRNA associated with seed UCCCUUU
| Name | miRBASE accession |
|---|---|
|
hsa-miR-204-5p
|
MIMAT0000265 |
|
hsa-miR-211-5p
|
MIMAT0000268 |
Activity profile of UCCCUUU motif
Sorted Z-values of UCCCUUU motif
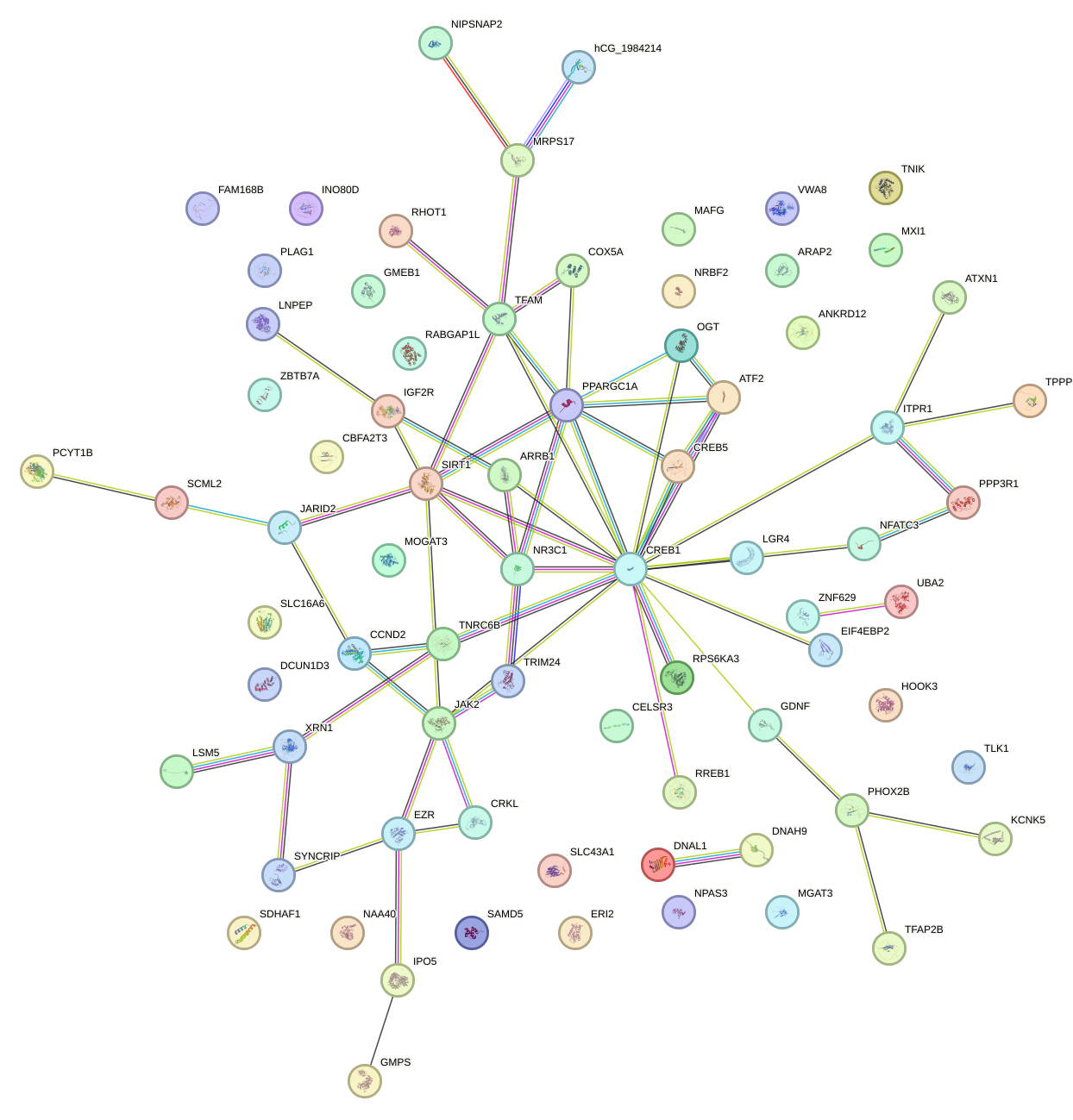
Network of associatons between targets according to the STRING database.
First level regulatory network of UCCCUUU
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_65613217 | 0.42 |
ENST00000545314.1 |
AK4 |
adenylate kinase 4 |
| chr6_+_15246501 | 0.37 |
ENST00000341776.2 |
JARID2 |
jumonji, AT rich interactive domain 2 |
| chr12_+_4382917 | 0.34 |
ENST00000261254.3 |
CCND2 |
cyclin D2 |
| chr17_+_55162453 | 0.34 |
ENST00000575322.1 ENST00000337714.3 ENST00000314126.3 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr3_-_138553594 | 0.33 |
ENST00000477593.1 ENST00000483968.1 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr6_+_160390102 | 0.31 |
ENST00000356956.1 |
IGF2R |
insulin-like growth factor 2 receptor |
| chr4_-_23891693 | 0.30 |
ENST00000264867.2 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr11_-_57283159 | 0.29 |
ENST00000533263.1 ENST00000278426.3 |
SLC43A1 |
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr10_+_88516396 | 0.29 |
ENST00000372037.3 |
BMPR1A |
bone morphogenetic protein receptor, type IA |
| chr7_+_138145076 | 0.27 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr16_-_89007491 | 0.23 |
ENST00000327483.5 ENST00000564416.1 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr2_-_68479614 | 0.22 |
ENST00000234310.3 |
PPP3R1 |
protein phosphatase 3, regulatory subunit B, alpha |
| chr11_+_63706444 | 0.22 |
ENST00000377793.4 ENST00000456907.2 ENST00000539656.1 |
NAA40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr8_+_104831472 | 0.21 |
ENST00000262231.10 ENST00000507740.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr6_+_10556215 | 0.20 |
ENST00000316170.3 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr19_+_36486078 | 0.20 |
ENST00000378887.2 |
SDHAF1 |
succinate dehydrogenase complex assembly factor 1 |
| chr6_-_159239257 | 0.20 |
ENST00000337147.7 ENST00000392177.4 |
EZR |
ezrin |
| chr17_-_66287257 | 0.19 |
ENST00000327268.4 |
SLC16A6 |
solute carrier family 16, member 6 |
| chr6_+_7107999 | 0.19 |
ENST00000491191.1 ENST00000379938.2 ENST00000471433.1 |
RREB1 |
ras responsive element binding protein 1 |
| chr19_+_34919257 | 0.19 |
ENST00000246548.4 ENST00000590048.2 |
UBA2 |
ubiquitin-like modifier activating enzyme 2 |
| chr4_-_36246060 | 0.19 |
ENST00000303965.4 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr16_-_30798492 | 0.18 |
ENST00000262525.4 |
ZNF629 |
zinc finger protein 629 |
| chr9_+_4985228 | 0.17 |
ENST00000381652.3 |
JAK2 |
Janus kinase 2 |
| chr17_-_79885576 | 0.16 |
ENST00000574686.1 ENST00000357736.4 |
MAFG |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr6_+_126112001 | 0.16 |
ENST00000392477.2 |
NCOA7 |
nuclear receptor coactivator 7 |
| chr2_-_200322723 | 0.16 |
ENST00000417098.1 |
SATB2 |
SATB homeobox 2 |
| chr8_+_42752053 | 0.16 |
ENST00000307602.4 |
HOOK3 |
hook microtubule-tethering protein 3 |
| chrX_-_110655391 | 0.16 |
ENST00000356915.2 ENST00000356220.3 |
DCX |
doublecortin |
| chr10_+_60144782 | 0.15 |
ENST00000487519.1 |
TFAM |
transcription factor A, mitochondrial |
| chr5_-_142783175 | 0.15 |
ENST00000231509.3 ENST00000394464.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr13_+_98605902 | 0.15 |
ENST00000460070.1 ENST00000481455.1 ENST00000261574.5 ENST00000493281.1 ENST00000463157.1 ENST00000471898.1 ENST00000489058.1 ENST00000481689.1 |
IPO5 |
importin 5 |
| chr19_-_4066890 | 0.14 |
ENST00000322357.4 |
ZBTB7A |
zinc finger and BTB domain containing 7A |
| chr16_-_20911641 | 0.14 |
ENST00000564349.1 ENST00000324344.4 |
ERI2 DCUN1D3 |
ERI1 exoribonuclease family member 2 DCN1, defective in cullin neddylation 1, domain containing 3 |
| chr22_+_40573921 | 0.14 |
ENST00000454349.2 ENST00000335727.9 |
TNRC6B |
trinucleotide repeat containing 6B |
| chr16_-_73082274 | 0.14 |
ENST00000268489.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr18_-_60987220 | 0.14 |
ENST00000398117.1 |
BCL2 |
B-cell CLL/lymphoma 2 |
| chr1_+_28995231 | 0.14 |
ENST00000373816.1 |
GMEB1 |
glucocorticoid modulatory element binding protein 1 |
| chrX_-_20284958 | 0.14 |
ENST00000379565.3 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr6_-_39197226 | 0.13 |
ENST00000359534.3 |
KCNK5 |
potassium channel, subfamily K, member 5 |
| chr22_+_21271714 | 0.13 |
ENST00000354336.3 |
CRKL |
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr6_-_16761678 | 0.12 |
ENST00000244769.4 ENST00000436367.1 |
ATXN1 |
ataxin 1 |
| chr12_+_51318513 | 0.12 |
ENST00000332160.4 |
METTL7A |
methyltransferase like 7A |
| chr15_-_34628951 | 0.12 |
ENST00000397707.2 ENST00000560611.1 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr8_-_57123815 | 0.11 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr5_-_693500 | 0.11 |
ENST00000360578.5 |
TPPP |
tubulin polymerization promoting protein |
| chr3_-_142166904 | 0.11 |
ENST00000264951.4 |
XRN1 |
5'-3' exoribonuclease 1 |
| chr6_+_50786414 | 0.11 |
ENST00000344788.3 ENST00000393655.3 ENST00000263046.4 |
TFAP2B |
transcription factor AP-2 beta (activating enhancer binding protein 2 beta) |
| chr3_+_4535025 | 0.11 |
ENST00000302640.8 ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr16_+_68119440 | 0.11 |
ENST00000346183.3 ENST00000329524.4 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chrX_-_18372792 | 0.11 |
ENST00000251900.4 |
SCML2 |
sex comb on midleg-like 2 (Drosophila) |
| chr11_-_27494279 | 0.10 |
ENST00000379214.4 |
LGR4 |
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr4_-_111119804 | 0.10 |
ENST00000394607.3 ENST00000302274.3 |
ELOVL6 |
ELOVL fatty acid elongase 6 |
| chrX_-_131352152 | 0.10 |
ENST00000342983.2 |
RAP2C |
RAP2C, member of RAS oncogene family |
| chr4_-_41750922 | 0.09 |
ENST00000226382.2 |
PHOX2B |
paired-like homeobox 2b |
| chr6_+_147830063 | 0.09 |
ENST00000367474.1 |
SAMD5 |
sterile alpha motif domain containing 5 |
| chr7_+_142552792 | 0.09 |
ENST00000392957.2 ENST00000442129.1 |
EPHB6 |
EPH receptor B6 |
| chr4_-_140098339 | 0.09 |
ENST00000394235.2 |
ELF2 |
E74-like factor 2 (ets domain transcription factor) |
| chr10_+_64893039 | 0.09 |
ENST00000277746.6 ENST00000435510.2 |
NRBF2 |
nuclear receptor binding factor 2 |
| chr13_-_42535214 | 0.09 |
ENST00000379310.3 ENST00000281496.6 |
VWA8 |
von Willebrand factor A domain containing 8 |
| chr5_+_96271141 | 0.09 |
ENST00000231368.5 |
LNPEP |
leucyl/cystinyl aminopeptidase |
| chr3_+_155588300 | 0.09 |
ENST00000496455.2 |
GMPS |
guanine monphosphate synthase |
| chr7_-_32529973 | 0.09 |
ENST00000410044.1 ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5 |
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr13_-_31736027 | 0.08 |
ENST00000380406.5 ENST00000320027.5 ENST00000380405.4 |
HSPH1 |
heat shock 105kDa/110kDa protein 1 |
| chr1_-_217262969 | 0.08 |
ENST00000361525.3 |
ESRRG |
estrogen-related receptor gamma |
| chr1_+_174769006 | 0.08 |
ENST00000489615.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr10_+_72164135 | 0.08 |
ENST00000373218.4 |
EIF4EBP2 |
eukaryotic translation initiation factor 4E binding protein 2 |
| chr15_-_75230478 | 0.08 |
ENST00000322347.6 |
COX5A |
cytochrome c oxidase subunit Va |
| chrX_+_28605516 | 0.08 |
ENST00000378993.1 |
IL1RAPL1 |
interleukin 1 receptor accessory protein-like 1 |
| chr2_-_206950781 | 0.08 |
ENST00000403263.1 |
INO80D |
INO80 complex subunit D |
| chr14_+_74111578 | 0.08 |
ENST00000554113.1 ENST00000555631.2 ENST00000553645.2 ENST00000311089.3 ENST00000555919.3 ENST00000554339.1 ENST00000554871.1 |
DNAL1 |
dynein, axonemal, light chain 1 |
| chr10_+_69644404 | 0.08 |
ENST00000212015.6 |
SIRT1 |
sirtuin 1 |
| chr3_-_171178157 | 0.08 |
ENST00000465393.1 ENST00000436636.2 ENST00000369326.5 ENST00000538048.1 ENST00000341852.6 |
TNIK |
TRAF2 and NCK interacting kinase |
| chrX_+_70752917 | 0.07 |
ENST00000373719.3 |
OGT |
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr4_-_66536057 | 0.07 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chr10_+_111967345 | 0.07 |
ENST00000332674.5 ENST00000453116.1 |
MXI1 |
MAX interactor 1, dimerization protein |
| chr22_+_39853258 | 0.07 |
ENST00000341184.6 |
MGAT3 |
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr19_+_10527449 | 0.07 |
ENST00000592685.1 ENST00000380702.2 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr7_+_28452130 | 0.07 |
ENST00000357727.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr11_-_75062730 | 0.07 |
ENST00000420843.2 ENST00000360025.3 |
ARRB1 |
arrestin, beta 1 |
| chr17_+_30469473 | 0.07 |
ENST00000333942.6 ENST00000358365.3 ENST00000583994.1 ENST00000545287.2 |
RHOT1 |
ras homolog family member T1 |
| chr7_+_56019486 | 0.07 |
ENST00000446692.1 ENST00000285298.4 ENST00000443449.1 |
GBAS MRPS17 |
glioblastoma amplified sequence mitochondrial ribosomal protein S17 |
| chr6_-_86352642 | 0.07 |
ENST00000355238.6 |
SYNCRIP |
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr2_-_176032843 | 0.07 |
ENST00000392544.1 ENST00000409499.1 ENST00000426833.3 ENST00000392543.2 ENST00000538946.1 ENST00000487334.2 ENST00000409833.1 ENST00000409635.1 ENST00000264110.2 ENST00000345739.5 |
ATF2 |
activating transcription factor 2 |
| chr12_+_69864129 | 0.06 |
ENST00000547219.1 ENST00000299293.2 ENST00000549921.1 ENST00000550316.1 ENST00000548154.1 ENST00000547414.1 ENST00000550389.1 ENST00000550937.1 ENST00000549092.1 ENST00000550169.1 |
FRS2 |
fibroblast growth factor receptor substrate 2 |
| chr1_-_9970227 | 0.06 |
ENST00000377263.1 |
CTNNBIP1 |
catenin, beta interacting protein 1 |
| chrX_-_24690771 | 0.06 |
ENST00000379145.1 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr8_+_77593448 | 0.06 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr2_-_172017343 | 0.06 |
ENST00000431350.2 ENST00000360843.3 |
TLK1 |
tousled-like kinase 1 |
| chr3_-_48700310 | 0.06 |
ENST00000164024.4 ENST00000544264.1 |
CELSR3 |
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr14_+_33408449 | 0.06 |
ENST00000346562.2 ENST00000341321.4 ENST00000548645.1 ENST00000356141.4 ENST00000357798.5 |
NPAS3 |
neuronal PAS domain protein 3 |
| chr18_+_9136758 | 0.06 |
ENST00000383440.2 ENST00000262126.4 ENST00000577992.1 |
ANKRD12 |
ankyrin repeat domain 12 |
| chr2_-_2334888 | 0.06 |
ENST00000428368.2 ENST00000399161.2 |
MYT1L |
myelin transcription factor 1-like |
| chr2_+_208394616 | 0.05 |
ENST00000432329.2 ENST00000353267.3 ENST00000445803.1 |
CREB1 |
cAMP responsive element binding protein 1 |
| chr2_-_131850951 | 0.05 |
ENST00000409185.1 ENST00000389915.3 |
FAM168B |
family with sequence similarity 168, member B |
| chr6_+_152011628 | 0.05 |
ENST00000404742.1 ENST00000440973.1 |
ESR1 |
estrogen receptor 1 |
| chr19_-_11591848 | 0.05 |
ENST00000359227.3 |
ELAVL3 |
ELAV like neuron-specific RNA binding protein 3 |
| chr4_+_99916765 | 0.05 |
ENST00000296411.6 |
METAP1 |
methionyl aminopeptidase 1 |
| chr19_-_38714847 | 0.05 |
ENST00000420980.2 ENST00000355526.4 |
DPF1 |
D4, zinc and double PHD fingers family 1 |
| chr1_+_36348790 | 0.05 |
ENST00000373204.4 |
AGO1 |
argonaute RISC catalytic component 1 |
| chr3_+_169940153 | 0.05 |
ENST00000295797.4 |
PRKCI |
protein kinase C, iota |
| chr12_+_20522179 | 0.05 |
ENST00000359062.3 |
PDE3A |
phosphodiesterase 3A, cGMP-inhibited |
| chr2_+_109335929 | 0.05 |
ENST00000283195.6 |
RANBP2 |
RAN binding protein 2 |
| chr1_-_70671216 | 0.05 |
ENST00000370952.3 |
LRRC40 |
leucine rich repeat containing 40 |
| chr2_-_157189180 | 0.05 |
ENST00000539077.1 ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
| chr2_-_152684977 | 0.05 |
ENST00000428992.2 ENST00000295087.8 |
ARL5A |
ADP-ribosylation factor-like 5A |
| chr3_+_183873098 | 0.05 |
ENST00000313143.3 |
DVL3 |
dishevelled segment polarity protein 3 |
| chr20_-_35374456 | 0.05 |
ENST00000373803.2 ENST00000359675.2 ENST00000540765.1 ENST00000349004.1 |
NDRG3 |
NDRG family member 3 |
| chr4_-_176923483 | 0.05 |
ENST00000280187.7 ENST00000512509.1 |
GPM6A |
glycoprotein M6A |
| chr9_-_20622478 | 0.04 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr12_+_57610562 | 0.04 |
ENST00000349394.5 |
NXPH4 |
neurexophilin 4 |
| chr5_-_95297678 | 0.04 |
ENST00000237853.4 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr19_-_14629224 | 0.04 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr14_-_99737565 | 0.04 |
ENST00000357195.3 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
| chr7_+_39989611 | 0.04 |
ENST00000181839.4 |
CDK13 |
cyclin-dependent kinase 13 |
| chr13_-_21476900 | 0.04 |
ENST00000400602.2 ENST00000255305.6 |
XPO4 |
exportin 4 |
| chr18_+_49866496 | 0.04 |
ENST00000442544.2 |
DCC |
deleted in colorectal carcinoma |
| chr19_-_49149553 | 0.04 |
ENST00000084798.4 |
CA11 |
carbonic anhydrase XI |
| chr16_+_81348528 | 0.04 |
ENST00000568107.2 |
GAN |
gigaxonin |
| chr7_-_127225620 | 0.04 |
ENST00000321407.2 |
GCC1 |
GRIP and coiled-coil domain containing 1 |
| chr11_+_35684288 | 0.04 |
ENST00000299413.5 |
TRIM44 |
tripartite motif containing 44 |
| chr4_-_153457197 | 0.04 |
ENST00000281708.4 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr6_-_8102714 | 0.04 |
ENST00000502429.1 ENST00000429723.2 ENST00000507463.1 ENST00000379715.5 |
EEF1E1 |
eukaryotic translation elongation factor 1 epsilon 1 |
| chr12_-_57030115 | 0.04 |
ENST00000379441.3 ENST00000179765.5 ENST00000551812.1 |
BAZ2A |
bromodomain adjacent to zinc finger domain, 2A |
| chr1_+_154377669 | 0.03 |
ENST00000368485.3 ENST00000344086.4 |
IL6R |
interleukin 6 receptor |
| chr15_-_27018175 | 0.03 |
ENST00000311550.5 |
GABRB3 |
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr22_-_42017021 | 0.03 |
ENST00000263256.6 |
DESI1 |
desumoylating isopeptidase 1 |
| chr4_-_89205879 | 0.03 |
ENST00000608933.1 ENST00000315194.4 ENST00000514204.1 |
PPM1K |
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr7_+_94285637 | 0.03 |
ENST00000482108.1 ENST00000488574.1 |
PEG10 |
paternally expressed 10 |
| chr3_-_52312636 | 0.03 |
ENST00000296490.3 |
WDR82 |
WD repeat domain 82 |
| chr15_+_57210818 | 0.03 |
ENST00000438423.2 ENST00000267811.5 ENST00000452095.2 ENST00000559609.1 ENST00000333725.5 |
TCF12 |
transcription factor 12 |
| chr7_+_145813453 | 0.03 |
ENST00000361727.3 |
CNTNAP2 |
contactin associated protein-like 2 |
| chr12_+_54402790 | 0.03 |
ENST00000040584.4 |
HOXC8 |
homeobox C8 |
| chr7_+_138916231 | 0.03 |
ENST00000473989.3 ENST00000288561.8 |
UBN2 |
ubinuclein 2 |
| chr3_+_152017181 | 0.03 |
ENST00000498502.1 ENST00000324196.5 ENST00000545754.1 ENST00000357472.3 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr1_-_6240183 | 0.03 |
ENST00000262450.3 ENST00000378021.1 |
CHD5 |
chromodomain helicase DNA binding protein 5 |
| chr12_-_111021110 | 0.03 |
ENST00000354300.3 |
PPTC7 |
PTC7 protein phosphatase homolog (S. cerevisiae) |
| chr3_-_120068143 | 0.03 |
ENST00000295628.3 |
LRRC58 |
leucine rich repeat containing 58 |
| chr1_-_22469459 | 0.03 |
ENST00000290167.6 |
WNT4 |
wingless-type MMTV integration site family, member 4 |
| chr3_-_100120223 | 0.02 |
ENST00000284320.5 |
TOMM70A |
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr7_-_127983877 | 0.02 |
ENST00000415472.2 ENST00000478061.1 ENST00000223073.2 ENST00000459726.1 |
RBM28 |
RNA binding motif protein 28 |
| chr2_+_96931834 | 0.02 |
ENST00000488633.1 |
CIAO1 |
cytosolic iron-sulfur protein assembly 1 |
| chr1_+_147013182 | 0.02 |
ENST00000234739.3 |
BCL9 |
B-cell CLL/lymphoma 9 |
| chr14_+_57735614 | 0.02 |
ENST00000261558.3 |
AP5M1 |
adaptor-related protein complex 5, mu 1 subunit |
| chr4_+_78978724 | 0.02 |
ENST00000325942.6 ENST00000264895.6 ENST00000264899.6 |
FRAS1 |
Fraser syndrome 1 |
| chr6_-_94129244 | 0.02 |
ENST00000369303.4 ENST00000369297.1 |
EPHA7 |
EPH receptor A7 |
| chr17_-_40021656 | 0.02 |
ENST00000319121.3 |
KLHL11 |
kelch-like family member 11 |
| chr7_-_158622210 | 0.02 |
ENST00000251527.5 |
ESYT2 |
extended synaptotagmin-like protein 2 |
| chr11_-_132813566 | 0.02 |
ENST00000331898.7 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
| chr16_+_67063036 | 0.02 |
ENST00000290858.6 ENST00000564034.1 |
CBFB |
core-binding factor, beta subunit |
| chr12_+_123868320 | 0.02 |
ENST00000402868.3 ENST00000330479.4 |
SETD8 |
SET domain containing (lysine methyltransferase) 8 |
| chr1_+_32930647 | 0.02 |
ENST00000609129.1 |
ZBTB8B |
zinc finger and BTB domain containing 8B |
| chr9_+_133454943 | 0.02 |
ENST00000319725.9 |
FUBP3 |
far upstream element (FUSE) binding protein 3 |
| chr11_-_66115032 | 0.02 |
ENST00000311181.4 |
B3GNT1 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 |
| chr5_-_37839782 | 0.02 |
ENST00000326524.2 ENST00000515058.1 |
GDNF |
glial cell derived neurotrophic factor |
| chr3_-_69171708 | 0.02 |
ENST00000420581.2 |
LMOD3 |
leiomodin 3 (fetal) |
| chr15_+_52121822 | 0.02 |
ENST00000558455.1 ENST00000308580.7 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
| chrX_-_111326004 | 0.02 |
ENST00000262839.2 |
TRPC5 |
transient receptor potential cation channel, subfamily C, member 5 |
| chr5_-_66492562 | 0.02 |
ENST00000256447.4 |
CD180 |
CD180 molecule |
| chr10_-_121356007 | 0.02 |
ENST00000369093.2 ENST00000436547.2 |
TIAL1 |
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr10_+_93683519 | 0.02 |
ENST00000265990.6 |
BTAF1 |
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa |
| chr20_+_20348740 | 0.01 |
ENST00000310227.1 |
INSM1 |
insulinoma-associated 1 |
| chr6_+_45389893 | 0.01 |
ENST00000371432.3 |
RUNX2 |
runt-related transcription factor 2 |
| chr4_-_8873531 | 0.01 |
ENST00000400677.3 |
HMX1 |
H6 family homeobox 1 |
| chr10_-_61666267 | 0.01 |
ENST00000263102.6 |
CCDC6 |
coiled-coil domain containing 6 |
| chr7_-_105029329 | 0.01 |
ENST00000393651.3 ENST00000460391.1 |
SRPK2 |
SRSF protein kinase 2 |
| chr7_+_100797678 | 0.01 |
ENST00000337619.5 |
AP1S1 |
adaptor-related protein complex 1, sigma 1 subunit |
| chr8_-_17270809 | 0.01 |
ENST00000180173.5 ENST00000521857.1 |
MTMR7 |
myotubularin related protein 7 |
| chr10_-_79686284 | 0.01 |
ENST00000372391.2 ENST00000372388.2 |
DLG5 |
discs, large homolog 5 (Drosophila) |
| chr18_+_32558208 | 0.01 |
ENST00000436190.2 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr10_+_12171721 | 0.01 |
ENST00000379020.4 ENST00000379017.3 |
SEC61A2 |
Sec61 alpha 2 subunit (S. cerevisiae) |
| chr4_-_38858428 | 0.01 |
ENST00000436693.2 ENST00000508254.1 ENST00000514655.1 ENST00000506146.1 |
TLR6 TLR1 |
toll-like receptor 6 toll-like receptor 1 |
| chr11_+_34073195 | 0.01 |
ENST00000341394.4 |
CAPRIN1 |
cell cycle associated protein 1 |
| chr11_+_111957497 | 0.01 |
ENST00000375549.3 ENST00000528182.1 ENST00000528048.1 ENST00000528021.1 ENST00000526592.1 ENST00000525291.1 |
SDHD |
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr13_-_45915221 | 0.01 |
ENST00000309246.5 ENST00000379060.4 ENST00000379055.1 ENST00000527226.1 ENST00000379056.1 |
TPT1 |
tumor protein, translationally-controlled 1 |
| chr10_-_81965307 | 0.01 |
ENST00000537102.1 ENST00000372231.3 ENST00000438331.1 ENST00000422982.3 ENST00000360615.4 ENST00000265447.4 |
ANXA11 |
annexin A11 |
| chr4_-_68566832 | 0.01 |
ENST00000420827.2 ENST00000322244.5 |
UBA6 |
ubiquitin-like modifier activating enzyme 6 |
| chr11_+_925824 | 0.01 |
ENST00000525796.1 ENST00000534328.1 ENST00000448903.2 ENST00000332231.5 |
AP2A2 |
adaptor-related protein complex 2, alpha 2 subunit |
| chr10_+_99344104 | 0.01 |
ENST00000555577.1 ENST00000370649.3 |
PI4K2A PI4K2A |
phosphatidylinositol 4-kinase type 2 alpha Phosphatidylinositol 4-kinase type 2-alpha; Uncharacterized protein |
| chr1_+_193091080 | 0.01 |
ENST00000367435.3 |
CDC73 |
cell division cycle 73 |
| chr19_+_10828724 | 0.00 |
ENST00000585892.1 ENST00000314646.5 ENST00000359692.6 |
DNM2 |
dynamin 2 |
| chr16_+_70148230 | 0.00 |
ENST00000398122.3 ENST00000568530.1 |
PDPR |
pyruvate dehydrogenase phosphatase regulatory subunit |
| chr13_-_28674693 | 0.00 |
ENST00000537084.1 ENST00000241453.7 ENST00000380982.4 |
FLT3 |
fms-related tyrosine kinase 3 |
| chr22_-_37099555 | 0.00 |
ENST00000300105.6 |
CACNG2 |
calcium channel, voltage-dependent, gamma subunit 2 |
| chr17_+_36508111 | 0.00 |
ENST00000331159.5 ENST00000577233.1 |
SOCS7 |
suppressor of cytokine signaling 7 |
| chr2_+_192542850 | 0.00 |
ENST00000410026.2 |
NABP1 |
nucleic acid binding protein 1 |
| chr3_-_9811595 | 0.00 |
ENST00000256460.3 |
CAMK1 |
calcium/calmodulin-dependent protein kinase I |
| chr3_+_88188254 | 0.00 |
ENST00000309495.5 |
ZNF654 |
zinc finger protein 654 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.2 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.0 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.3 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.1 | 0.1 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.1 | 0.4 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 0.3 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.2 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.2 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.2 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) terminal web assembly(GO:1902896) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0046671 | negative regulation of cellular pH reduction(GO:0032848) CD8-positive, alpha-beta T cell lineage commitment(GO:0043375) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.1 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.4 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0061290 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 0.0 | 0.2 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.2 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.1 | GO:0072501 | cellular divalent inorganic anion homeostasis(GO:0072501) |
| 0.0 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.0 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.1 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) |
| 0.0 | 0.2 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:1903750 | negative regulation of establishment of protein localization to mitochondrion(GO:1903748) regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.2 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.0 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.2 | GO:0016018 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |