Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for VAX1_GSX2
Z-value: 1.06
Transcription factors associated with VAX1_GSX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VAX1
|
ENSG00000148704.8 | VAX1 |
|
GSX2
|
ENSG00000180613.6 | GSX2 |
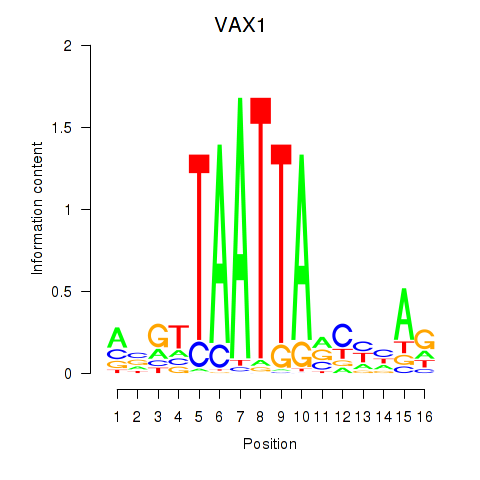
Activity profile of VAX1_GSX2 motif
Sorted Z-values of VAX1_GSX2 motif
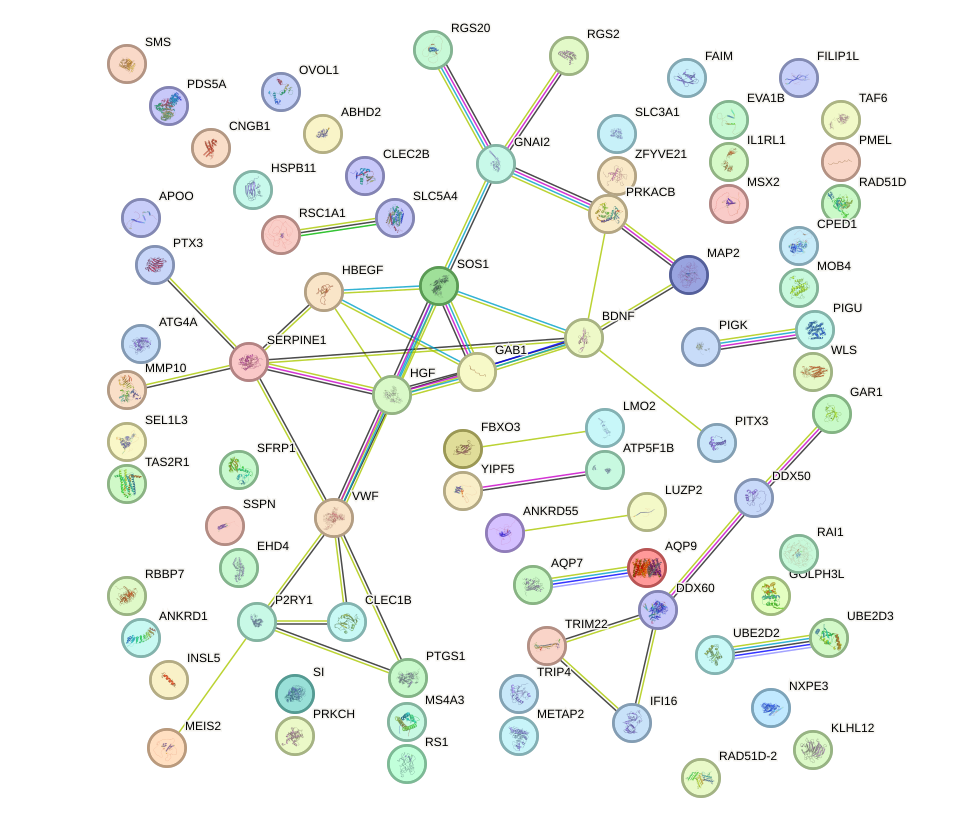
Network of associatons between targets according to the STRING database.
First level regulatory network of VAX1_GSX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_125133315 | 2.72 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr9_+_125132803 | 2.50 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr3_+_157154578 | 2.48 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr1_+_192778161 | 2.36 |
ENST00000235382.5 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
| chr1_-_68698222 | 2.31 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr7_-_81399329 | 2.15 |
ENST00000453411.1 ENST00000444829.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr7_-_81399438 | 1.95 |
ENST00000222390.5 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr8_-_41166953 | 1.75 |
ENST00000220772.3 |
SFRP1 |
secreted frizzled-related protein 1 |
| chr4_-_25865159 | 1.66 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr12_-_10022735 | 1.54 |
ENST00000228438.2 |
CLEC2B |
C-type lectin domain family 2, member B |
| chr1_-_68698197 | 1.53 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr12_-_6233828 | 1.30 |
ENST00000572068.1 ENST00000261405.5 |
VWF |
von Willebrand factor |
| chr7_-_81399411 | 1.21 |
ENST00000423064.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr3_+_62304712 | 1.17 |
ENST00000494481.1 |
C3orf14 |
chromosome 3 open reading frame 14 |
| chr12_+_26348246 | 1.15 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr7_+_100770328 | 1.13 |
ENST00000223095.4 ENST00000445463.2 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr12_+_29376673 | 1.10 |
ENST00000547116.1 |
FAR2 |
fatty acyl CoA reductase 2 |
| chr11_-_27722021 | 1.09 |
ENST00000356660.4 ENST00000418212.1 ENST00000533246.1 |
BDNF |
brain-derived neurotrophic factor |
| chr12_+_29376592 | 1.09 |
ENST00000182377.4 |
FAR2 |
fatty acyl CoA reductase 2 |
| chr22_-_32651326 | 1.09 |
ENST00000266086.4 |
SLC5A4 |
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr3_+_62304648 | 1.07 |
ENST00000462069.1 ENST00000232519.5 ENST00000465142.1 |
C3orf14 |
chromosome 3 open reading frame 14 |
| chr1_-_36789755 | 0.99 |
ENST00000270824.1 |
EVA1B |
eva-1 homolog B (C. elegans) |
| chr3_+_50273625 | 0.99 |
ENST00000536647.1 |
GNAI2 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr7_-_81399355 | 0.97 |
ENST00000457544.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr15_-_37393406 | 0.92 |
ENST00000338564.5 ENST00000558313.1 ENST00000340545.5 |
MEIS2 |
Meis homeobox 2 |
| chr4_-_169239921 | 0.92 |
ENST00000514995.1 ENST00000393743.3 |
DDX60 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr7_+_120628731 | 0.84 |
ENST00000310396.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr2_+_102953608 | 0.84 |
ENST00000311734.2 ENST00000409584.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chr7_-_99716952 | 0.84 |
ENST00000523306.1 ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr1_+_158978768 | 0.83 |
ENST00000447473.2 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr1_+_81771806 | 0.80 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr11_+_5710919 | 0.72 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr3_+_158519654 | 0.68 |
ENST00000415822.2 ENST00000392813.4 ENST00000264266.8 |
MFSD1 |
major facilitator superfamily domain containing 1 |
| chr5_-_143550159 | 0.66 |
ENST00000448443.2 ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5 |
Yip1 domain family, member 5 |
| chr11_-_33774944 | 0.63 |
ENST00000532057.1 ENST00000531080.1 |
FBXO3 |
F-box protein 3 |
| chr2_+_44502597 | 0.62 |
ENST00000260649.6 ENST00000409387.1 |
SLC3A1 |
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr15_+_64680003 | 0.62 |
ENST00000261884.3 |
TRIP4 |
thyroid hormone receptor interactor 4 |
| chr11_-_33913708 | 0.61 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr14_+_61654271 | 0.60 |
ENST00000555185.1 ENST00000557294.1 ENST00000556778.1 |
PRKCH |
protein kinase C, eta |
| chr1_-_202897724 | 0.59 |
ENST00000435533.3 ENST00000367258.1 |
KLHL12 |
kelch-like family member 12 |
| chr14_+_104182105 | 0.59 |
ENST00000311141.2 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr14_+_104182061 | 0.58 |
ENST00000216602.6 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr11_+_24518723 | 0.56 |
ENST00000336930.6 ENST00000529015.1 ENST00000533227.1 |
LUZP2 |
leucine zipper protein 2 |
| chr4_-_164534657 | 0.56 |
ENST00000339875.5 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr1_+_84630645 | 0.55 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr15_-_55563072 | 0.54 |
ENST00000567380.1 ENST00000565972.1 ENST00000569493.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr1_-_151965048 | 0.52 |
ENST00000368809.1 |
S100A10 |
S100 calcium binding protein A10 |
| chr17_-_66951474 | 0.51 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr7_-_14029283 | 0.51 |
ENST00000433547.1 ENST00000405192.2 |
ETV1 |
ets variant 1 |
| chr12_+_26348429 | 0.51 |
ENST00000242729.2 |
SSPN |
sarcospan |
| chr5_-_55412774 | 0.50 |
ENST00000434982.2 |
ANKRD55 |
ankyrin repeat domain 55 |
| chr11_-_102651343 | 0.50 |
ENST00000279441.4 ENST00000539681.1 |
MMP10 |
matrix metallopeptidase 10 (stromelysin 2) |
| chrX_+_107334895 | 0.50 |
ENST00000372232.3 ENST00000345734.3 ENST00000372254.3 |
ATG4A |
autophagy related 4A, cysteine peptidase |
| chr15_-_55562582 | 0.49 |
ENST00000396307.2 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr5_-_139726181 | 0.47 |
ENST00000507104.1 ENST00000230990.6 |
HBEGF |
heparin-binding EGF-like growth factor |
| chr15_+_84115868 | 0.46 |
ENST00000427482.2 |
SH3GL3 |
SH3-domain GRB2-like 3 |
| chrX_-_18690210 | 0.45 |
ENST00000379984.3 |
RS1 |
retinoschisin 1 |
| chr12_-_57039739 | 0.43 |
ENST00000552959.1 ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B |
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chrX_-_23926004 | 0.41 |
ENST00000379226.4 ENST00000379220.3 |
APOO |
apolipoprotein O |
| chr8_+_54764346 | 0.41 |
ENST00000297313.3 ENST00000344277.6 |
RGS20 |
regulator of G-protein signaling 20 |
| chr1_-_77685084 | 0.40 |
ENST00000370812.3 ENST00000359130.1 ENST00000445065.1 ENST00000370813.5 |
PIGK |
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr2_+_210517895 | 0.40 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr15_-_42264702 | 0.39 |
ENST00000220325.4 |
EHD4 |
EH-domain containing 4 |
| chr15_-_55562479 | 0.38 |
ENST00000564609.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr11_+_59824060 | 0.38 |
ENST00000395032.2 ENST00000358152.2 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr17_-_33446735 | 0.38 |
ENST00000460118.2 ENST00000335858.7 |
RAD51D |
RAD51 paralog D |
| chr3_+_101504200 | 0.36 |
ENST00000422132.1 |
NXPE3 |
neurexophilin and PC-esterase domain family, member 3 |
| chr11_+_59824127 | 0.35 |
ENST00000278865.3 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr3_-_99569821 | 0.35 |
ENST00000487087.1 |
FILIP1L |
filamin A interacting protein 1-like |
| chr1_-_67266939 | 0.32 |
ENST00000304526.2 |
INSL5 |
insulin-like 5 |
| chr4_+_144354644 | 0.31 |
ENST00000512843.1 |
GAB1 |
GRB2-associated binding protein 1 |
| chr10_-_92681033 | 0.31 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr11_-_71823266 | 0.30 |
ENST00000538919.1 ENST00000539395.1 ENST00000542531.1 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr6_-_110501200 | 0.30 |
ENST00000392586.1 ENST00000419252.1 ENST00000392589.1 ENST00000392588.1 ENST00000359451.2 |
WASF1 |
WAS protein family, member 1 |
| chr10_+_47894023 | 0.29 |
ENST00000358474.5 |
FAM21B |
family with sequence similarity 21, member B |
| chrX_+_21958674 | 0.28 |
ENST00000404933.2 |
SMS |
spermine synthase |
| chr11_+_65554493 | 0.28 |
ENST00000335987.3 |
OVOL1 |
ovo-like zinc finger 1 |
| chr1_-_150669500 | 0.28 |
ENST00000271732.3 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr2_+_198380289 | 0.27 |
ENST00000233892.4 ENST00000409916.1 |
MOB4 |
MOB family member 4, phocein |
| chr5_-_9630463 | 0.27 |
ENST00000382492.2 |
TAS2R1 |
taste receptor, type 2, member 1 |
| chr3_-_164796269 | 0.26 |
ENST00000264382.3 |
SI |
sucrase-isomaltase (alpha-glucosidase) |
| chr15_+_89631381 | 0.25 |
ENST00000352732.5 |
ABHD2 |
abhydrolase domain containing 2 |
| chr4_+_110736659 | 0.24 |
ENST00000394631.3 ENST00000226796.6 |
GAR1 |
GAR1 ribonucleoprotein |
| chr9_-_95166884 | 0.24 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr21_-_15918618 | 0.23 |
ENST00000400564.1 ENST00000400566.1 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr3_+_138340067 | 0.23 |
ENST00000479848.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr6_+_34204642 | 0.23 |
ENST00000347617.6 ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1 |
high mobility group AT-hook 1 |
| chr15_+_58430567 | 0.23 |
ENST00000536493.1 |
AQP9 |
aquaporin 9 |
| chr12_+_28410128 | 0.23 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr10_+_70661014 | 0.22 |
ENST00000373585.3 |
DDX50 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr2_-_44588624 | 0.22 |
ENST00000438314.1 ENST00000409936.1 |
PREPL |
prolyl endopeptidase-like |
| chrX_-_16887963 | 0.22 |
ENST00000380084.4 |
RBBP7 |
retinoblastoma binding protein 7 |
| chr1_-_54411255 | 0.22 |
ENST00000371377.3 |
HSPB11 |
heat shock protein family B (small), member 11 |
| chr5_-_94890648 | 0.22 |
ENST00000513823.1 ENST00000514952.1 ENST00000358746.2 |
TTC37 |
tetratricopeptide repeat domain 37 |
| chr7_+_77428149 | 0.21 |
ENST00000415251.2 ENST00000275575.7 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chr4_-_103749313 | 0.21 |
ENST00000394803.5 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr3_+_152552685 | 0.21 |
ENST00000305097.3 |
P2RY1 |
purinergic receptor P2Y, G-protein coupled, 1 |
| chr4_-_103749179 | 0.21 |
ENST00000502690.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr15_+_89631647 | 0.21 |
ENST00000569550.1 ENST00000565066.1 ENST00000565973.1 |
ABHD2 |
abhydrolase domain containing 2 |
| chr2_-_44588679 | 0.20 |
ENST00000409411.1 |
PREPL |
prolyl endopeptidase-like |
| chr5_+_174151536 | 0.20 |
ENST00000239243.6 ENST00000507785.1 |
MSX2 |
msh homeobox 2 |
| chr8_-_86290333 | 0.20 |
ENST00000521846.1 ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1 |
carbonic anhydrase I |
| chr4_-_103749205 | 0.20 |
ENST00000508249.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr11_-_71823796 | 0.20 |
ENST00000545680.1 ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr4_-_39979576 | 0.20 |
ENST00000303538.8 ENST00000503396.1 |
PDS5A |
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr21_-_35899113 | 0.19 |
ENST00000492600.1 ENST00000481448.1 ENST00000381132.2 |
RCAN1 |
regulator of calcineurin 1 |
| chr12_-_71031220 | 0.19 |
ENST00000334414.6 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr7_-_14028488 | 0.18 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr12_+_20963647 | 0.18 |
ENST00000381545.3 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr2_-_44588694 | 0.18 |
ENST00000409957.1 |
PREPL |
prolyl endopeptidase-like |
| chr7_-_99717463 | 0.18 |
ENST00000437822.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr8_-_86253888 | 0.18 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr19_+_19303008 | 0.18 |
ENST00000353145.1 ENST00000421262.3 ENST00000303088.4 ENST00000456252.3 ENST00000593273.1 |
RFXANK |
regulatory factor X-associated ankyrin-containing protein |
| chr4_-_103749105 | 0.18 |
ENST00000394801.4 ENST00000394804.2 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr13_-_36050819 | 0.17 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr7_+_77428066 | 0.17 |
ENST00000422959.2 ENST00000307305.8 ENST00000424760.1 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chr4_+_70796784 | 0.17 |
ENST00000246891.4 ENST00000444405.3 |
CSN1S1 |
casein alpha s1 |
| chr14_-_70883708 | 0.17 |
ENST00000256366.4 |
SYNJ2BP |
synaptojanin 2 binding protein |
| chr9_-_95166841 | 0.16 |
ENST00000262551.4 |
OGN |
osteoglycin |
| chr17_-_64187973 | 0.16 |
ENST00000583358.1 ENST00000392769.2 |
CEP112 |
centrosomal protein 112kDa |
| chr12_+_20963632 | 0.16 |
ENST00000540853.1 ENST00000261196.2 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr17_-_46690839 | 0.16 |
ENST00000498634.2 |
HOXB8 |
homeobox B8 |
| chr17_+_48823975 | 0.16 |
ENST00000513969.1 ENST00000503728.1 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
| chr1_-_109618566 | 0.15 |
ENST00000338366.5 |
TAF13 |
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr22_+_23154239 | 0.15 |
ENST00000390315.2 |
IGLV3-10 |
immunoglobulin lambda variable 3-10 |
| chr11_+_66276550 | 0.15 |
ENST00000419755.3 |
CTD-3074O7.11 |
Bardet-Biedl syndrome 1 protein |
| chr16_-_66764119 | 0.15 |
ENST00000569320.1 |
DYNC1LI2 |
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr11_-_71823715 | 0.15 |
ENST00000545944.1 ENST00000502597.2 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr8_-_62602327 | 0.15 |
ENST00000445642.3 ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH |
aspartate beta-hydroxylase |
| chr1_-_197115818 | 0.14 |
ENST00000367409.4 ENST00000294732.7 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr3_+_130569429 | 0.14 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr6_-_38670897 | 0.14 |
ENST00000373365.4 |
GLO1 |
glyoxalase I |
| chr12_-_10978957 | 0.14 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr6_+_26440700 | 0.14 |
ENST00000494393.1 ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
| chr6_+_29429217 | 0.13 |
ENST00000396792.2 |
OR2H1 |
olfactory receptor, family 2, subfamily H, member 1 |
| chr12_-_10959892 | 0.13 |
ENST00000240615.2 |
TAS2R8 |
taste receptor, type 2, member 8 |
| chr5_+_176811431 | 0.13 |
ENST00000512593.1 ENST00000324417.5 |
SLC34A1 |
solute carrier family 34 (type II sodium/phosphate contransporter), member 1 |
| chr1_+_16370271 | 0.12 |
ENST00000375679.4 |
CLCNKB |
chloride channel, voltage-sensitive Kb |
| chr5_+_140579162 | 0.11 |
ENST00000536699.1 ENST00000354757.3 |
PCDHB11 |
protocadherin beta 11 |
| chr21_-_43346790 | 0.11 |
ENST00000329623.7 |
C2CD2 |
C2 calcium-dependent domain containing 2 |
| chr3_+_149191723 | 0.11 |
ENST00000305354.4 |
TM4SF4 |
transmembrane 4 L six family member 4 |
| chr19_+_48949030 | 0.11 |
ENST00000253237.5 |
GRWD1 |
glutamate-rich WD repeat containing 1 |
| chr3_+_138340049 | 0.11 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr10_-_28571015 | 0.11 |
ENST00000375719.3 ENST00000375732.1 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr11_+_46958248 | 0.10 |
ENST00000536126.1 ENST00000278460.7 ENST00000378618.2 ENST00000395460.2 ENST00000378615.3 ENST00000543718.1 |
C11orf49 |
chromosome 11 open reading frame 49 |
| chr4_-_103748880 | 0.10 |
ENST00000453744.2 ENST00000349311.8 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr10_+_96443378 | 0.10 |
ENST00000285979.6 |
CYP2C18 |
cytochrome P450, family 2, subfamily C, polypeptide 18 |
| chr18_-_54305658 | 0.09 |
ENST00000586262.1 ENST00000217515.6 |
TXNL1 |
thioredoxin-like 1 |
| chr1_+_160160283 | 0.09 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr3_-_141747950 | 0.08 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_+_160160346 | 0.08 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr19_-_58951496 | 0.08 |
ENST00000254166.3 |
ZNF132 |
zinc finger protein 132 |
| chr7_+_55433131 | 0.08 |
ENST00000254770.2 |
LANCL2 |
LanC lantibiotic synthetase component C-like 2 (bacterial) |
| chr1_+_12834984 | 0.08 |
ENST00000357726.4 |
PRAMEF12 |
PRAME family member 12 |
| chr12_+_81110684 | 0.08 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr2_+_11682790 | 0.07 |
ENST00000389825.3 ENST00000381483.2 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr2_-_3521518 | 0.07 |
ENST00000382093.5 |
ADI1 |
acireductone dioxygenase 1 |
| chr1_-_207226313 | 0.07 |
ENST00000367084.1 |
YOD1 |
YOD1 deubiquitinase |
| chr11_-_13517565 | 0.06 |
ENST00000282091.1 ENST00000529816.1 |
PTH |
parathyroid hormone |
| chr2_+_169757750 | 0.06 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
| chr3_+_111717511 | 0.05 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr6_+_110501344 | 0.05 |
ENST00000368932.1 |
CDC40 |
cell division cycle 40 |
| chr17_-_2996290 | 0.05 |
ENST00000331459.1 |
OR1D2 |
olfactory receptor, family 1, subfamily D, member 2 |
| chrX_-_13835147 | 0.05 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr4_+_41614909 | 0.05 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr11_+_33061543 | 0.05 |
ENST00000432887.1 ENST00000528898.1 ENST00000531632.2 |
TCP11L1 |
t-complex 11, testis-specific-like 1 |
| chr12_-_10605929 | 0.04 |
ENST00000347831.5 ENST00000359151.3 |
KLRC1 |
killer cell lectin-like receptor subfamily C, member 1 |
| chr19_+_50016610 | 0.04 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr18_+_46065393 | 0.04 |
ENST00000256413.3 |
CTIF |
CBP80/20-dependent translation initiation factor |
| chr1_-_190446759 | 0.04 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr17_-_45266542 | 0.04 |
ENST00000531206.1 ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27 |
cell division cycle 27 |
| chr5_+_66300446 | 0.04 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr20_-_50722183 | 0.04 |
ENST00000371523.4 |
ZFP64 |
ZFP64 zinc finger protein |
| chr8_+_9413410 | 0.04 |
ENST00000520408.1 ENST00000310430.6 ENST00000522110.1 |
TNKS |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr10_-_61900762 | 0.04 |
ENST00000355288.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr1_-_205325850 | 0.04 |
ENST00000537168.1 |
KLHDC8A |
kelch domain containing 8A |
| chrX_-_77225135 | 0.03 |
ENST00000458128.1 |
PGAM4 |
phosphoglycerate mutase family member 4 |
| chr15_-_64665911 | 0.03 |
ENST00000606793.1 ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1 |
Uncharacterized protein |
| chr12_-_10607084 | 0.03 |
ENST00000408006.3 ENST00000544822.1 ENST00000536188.1 |
KLRC1 |
killer cell lectin-like receptor subfamily C, member 1 |
| chr3_+_186353756 | 0.03 |
ENST00000431018.1 ENST00000450521.1 ENST00000539949.1 |
FETUB |
fetuin B |
| chr17_-_10372875 | 0.03 |
ENST00000255381.2 |
MYH4 |
myosin, heavy chain 4, skeletal muscle |
| chr15_-_65903407 | 0.03 |
ENST00000395644.4 ENST00000567744.1 ENST00000568573.1 ENST00000562830.1 ENST00000569491.1 ENST00000561769.1 |
VWA9 |
von Willebrand factor A domain containing 9 |
| chr16_+_69345243 | 0.03 |
ENST00000254950.11 |
VPS4A |
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr1_-_1710229 | 0.03 |
ENST00000341991.3 |
NADK |
NAD kinase |
| chr2_-_169887827 | 0.03 |
ENST00000263817.6 |
ABCB11 |
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr1_+_186369704 | 0.02 |
ENST00000574641.1 |
OCLM |
oculomedin |
| chr17_-_3030875 | 0.02 |
ENST00000328890.2 |
OR1G1 |
olfactory receptor, family 1, subfamily G, member 1 |
| chr1_+_117544366 | 0.02 |
ENST00000256652.4 ENST00000369470.1 |
CD101 |
CD101 molecule |
| chr6_+_110501621 | 0.02 |
ENST00000368930.1 ENST00000307731.1 |
CDC40 |
cell division cycle 40 |
| chr16_+_6069072 | 0.02 |
ENST00000547605.1 ENST00000550418.1 ENST00000553186.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr18_+_22040620 | 0.02 |
ENST00000426880.2 |
HRH4 |
histamine receptor H4 |
| chr12_+_106751436 | 0.02 |
ENST00000228347.4 |
POLR3B |
polymerase (RNA) III (DNA directed) polypeptide B |
| chr15_+_62853562 | 0.01 |
ENST00000561311.1 |
TLN2 |
talin 2 |
| chr2_-_208031943 | 0.01 |
ENST00000421199.1 ENST00000457962.1 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr14_-_104181771 | 0.01 |
ENST00000554913.1 ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3 |
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr18_-_19997878 | 0.01 |
ENST00000391403.2 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
| chr6_+_28317685 | 0.01 |
ENST00000252211.2 ENST00000341464.5 ENST00000377255.3 |
ZKSCAN3 |
zinc finger with KRAB and SCAN domains 3 |
| chr8_-_97273807 | 0.01 |
ENST00000517720.1 ENST00000287025.3 ENST00000523821.1 |
MTERFD1 |
MTERF domain containing 1 |
| chr12_-_10955226 | 0.01 |
ENST00000240687.2 |
TAS2R7 |
taste receptor, type 2, member 7 |
| chr16_-_1464688 | 0.01 |
ENST00000389221.4 ENST00000508903.2 ENST00000397462.1 ENST00000301712.5 |
UNKL |
unkempt family zinc finger-like |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 5.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.1 | 1.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 1.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.9 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 2.2 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.8 | 6.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.7 | 2.2 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.6 | 1.7 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.5 | 2.5 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.4 | 1.1 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.4 | 1.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.3 | 2.4 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.3 | 5.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.2 | 1.4 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 0.9 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 0.5 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.1 | 0.8 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.5 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.1 | 0.6 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.1 | 0.6 | GO:1990822 | L-cystine transport(GO:0015811) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.3 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.3 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 1.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 0.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.2 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.1 | 0.4 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.2 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.4 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.1 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.6 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 1.1 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.5 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.3 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.6 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.2 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.5 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.0 | 0.7 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.6 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.5 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 1.2 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.0 | 0.1 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.2 | GO:0070884 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.4 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.3 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.0 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.0 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.0 | 0.6 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.1 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 1.2 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.7 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.6 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.3 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 3.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 2.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.4 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 7.4 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 1.0 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 5.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0090661 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 1.0 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.8 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.7 | 2.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.4 | 1.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.3 | 2.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 3.8 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.3 | 0.8 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.3 | 1.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 1.2 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.1 | 5.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.6 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.6 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 2.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.1 | 1.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 1.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 1.0 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |