Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for VDR
Z-value: 0.59
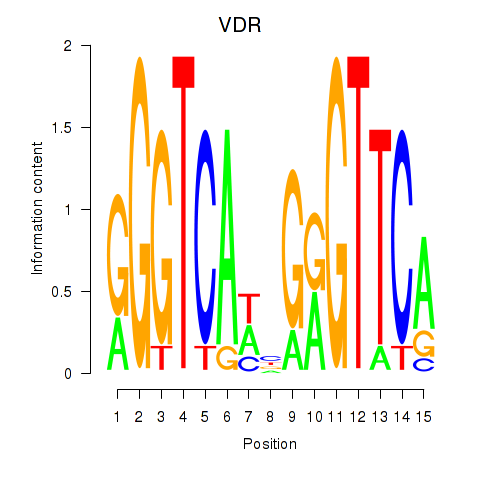
Transcription factors associated with VDR
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VDR
|
ENSG00000111424.6 | VDR |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VDR | hg19_v2_chr12_-_48298785_48298828 | 0.23 | 4.0e-01 | Click! |
Activity profile of VDR motif
Sorted Z-values of VDR motif
Network of associatons between targets according to the STRING database.
First level regulatory network of VDR
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_52626898 | 1.13 |
ENST00000331817.5 |
KRT7 |
keratin 7 |
| chr2_-_235405168 | 0.93 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr8_+_26371763 | 0.87 |
ENST00000521913.1 |
DPYSL2 |
dihydropyrimidinase-like 2 |
| chr17_-_7082861 | 0.84 |
ENST00000269299.3 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr2_+_11752379 | 0.84 |
ENST00000396123.1 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr17_+_2699697 | 0.83 |
ENST00000254695.8 ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
| chr8_+_95653427 | 0.82 |
ENST00000454170.2 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr8_+_95653302 | 0.79 |
ENST00000423620.2 ENST00000433389.2 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr17_-_64225508 | 0.78 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr8_+_95653373 | 0.78 |
ENST00000358397.5 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr4_+_85504075 | 0.75 |
ENST00000295887.5 |
CDS1 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr14_+_95047744 | 0.74 |
ENST00000553511.1 ENST00000554633.1 ENST00000555681.1 ENST00000554276.1 |
SERPINA5 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chrX_+_43515467 | 0.73 |
ENST00000338702.3 ENST00000542639.1 |
MAOA |
monoamine oxidase A |
| chr11_-_104817919 | 0.72 |
ENST00000533252.1 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr10_+_54074033 | 0.71 |
ENST00000373970.3 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
| chr9_+_71789133 | 0.66 |
ENST00000348208.4 ENST00000265384.7 |
TJP2 |
tight junction protein 2 |
| chr6_+_31916733 | 0.60 |
ENST00000483004.1 |
CFB |
complement factor B |
| chr7_+_94536898 | 0.53 |
ENST00000433360.1 ENST00000340694.4 ENST00000424654.1 |
PPP1R9A |
protein phosphatase 1, regulatory subunit 9A |
| chr19_+_41281416 | 0.51 |
ENST00000597140.1 |
MIA |
melanoma inhibitory activity |
| chr11_-_119991589 | 0.48 |
ENST00000526881.1 |
TRIM29 |
tripartite motif containing 29 |
| chr19_+_41281282 | 0.46 |
ENST00000263369.3 |
MIA |
melanoma inhibitory activity |
| chr8_-_22926623 | 0.43 |
ENST00000276431.4 |
TNFRSF10B |
tumor necrosis factor receptor superfamily, member 10b |
| chr19_+_41281060 | 0.43 |
ENST00000594436.1 ENST00000597784.1 |
MIA |
melanoma inhibitory activity |
| chr12_-_133050726 | 0.41 |
ENST00000595994.1 |
MUC8 |
mucin 8 |
| chrX_-_130423200 | 0.39 |
ENST00000361420.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr11_-_104840093 | 0.39 |
ENST00000417440.2 ENST00000444739.2 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr14_-_35873856 | 0.38 |
ENST00000553342.1 ENST00000216797.5 ENST00000557140.1 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr10_-_13523073 | 0.37 |
ENST00000440282.1 |
BEND7 |
BEN domain containing 7 |
| chr2_+_108905325 | 0.35 |
ENST00000438339.1 ENST00000409880.1 ENST00000437390.2 |
SULT1C2 |
sulfotransferase family, cytosolic, 1C, member 2 |
| chr7_-_95025661 | 0.32 |
ENST00000542556.1 ENST00000265627.5 ENST00000427422.1 ENST00000451904.1 |
PON1 PON3 |
paraoxonase 1 paraoxonase 3 |
| chr3_+_54157480 | 0.32 |
ENST00000490478.1 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr11_+_57365150 | 0.30 |
ENST00000457869.1 ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1 |
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr20_+_3801162 | 0.29 |
ENST00000379573.2 ENST00000379567.2 ENST00000455742.1 ENST00000246041.2 |
AP5S1 |
adaptor-related protein complex 5, sigma 1 subunit |
| chrX_-_13835147 | 0.28 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr19_+_54694119 | 0.27 |
ENST00000456872.1 ENST00000302937.4 ENST00000429671.2 |
TSEN34 |
TSEN34 tRNA splicing endonuclease subunit |
| chr10_-_33623826 | 0.26 |
ENST00000374867.2 |
NRP1 |
neuropilin 1 |
| chr2_-_203736334 | 0.26 |
ENST00000392237.2 ENST00000416760.1 ENST00000412210.1 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
| chr7_+_29234101 | 0.26 |
ENST00000435288.2 |
CHN2 |
chimerin 2 |
| chr2_+_210517895 | 0.25 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr2_+_54342574 | 0.25 |
ENST00000303536.4 ENST00000394666.3 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr8_-_145028013 | 0.24 |
ENST00000354958.2 |
PLEC |
plectin |
| chr19_+_1407517 | 0.24 |
ENST00000336761.6 ENST00000233078.4 |
DAZAP1 |
DAZ associated protein 1 |
| chr8_+_17434689 | 0.24 |
ENST00000398074.3 |
PDGFRL |
platelet-derived growth factor receptor-like |
| chr6_+_167704838 | 0.24 |
ENST00000366829.2 |
UNC93A |
unc-93 homolog A (C. elegans) |
| chr17_-_36347835 | 0.24 |
ENST00000519532.1 |
TBC1D3 |
TBC1 domain family, member 3 |
| chr21_-_47575481 | 0.24 |
ENST00000291670.5 ENST00000397748.1 ENST00000359679.2 ENST00000355384.2 ENST00000397746.3 ENST00000397743.1 |
FTCD |
formimidoyltransferase cyclodeaminase |
| chr16_+_19078960 | 0.24 |
ENST00000568985.1 ENST00000566110.1 |
COQ7 |
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr2_+_108905095 | 0.24 |
ENST00000251481.6 ENST00000326853.5 |
SULT1C2 |
sulfotransferase family, cytosolic, 1C, member 2 |
| chrX_-_13835461 | 0.23 |
ENST00000316715.4 ENST00000356942.5 |
GPM6B |
glycoprotein M6B |
| chr17_-_34756219 | 0.23 |
ENST00000451448.2 ENST00000394359.3 |
TBC1D3C TBC1D3H |
TBC1 domain family, member 3C TBC1 domain family, member 3H |
| chr6_+_30852130 | 0.23 |
ENST00000428153.2 ENST00000376568.3 ENST00000452441.1 ENST00000515219.1 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr6_+_167704798 | 0.23 |
ENST00000230256.3 |
UNC93A |
unc-93 homolog A (C. elegans) |
| chr8_-_22926526 | 0.22 |
ENST00000347739.3 ENST00000542226.1 |
TNFRSF10B |
tumor necrosis factor receptor superfamily, member 10b |
| chr8_-_13134045 | 0.22 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr16_+_19079311 | 0.21 |
ENST00000569127.1 |
COQ7 |
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr17_+_36284791 | 0.21 |
ENST00000505415.1 |
TBC1D3F |
TBC1 domain family, member 3F |
| chr17_-_36348610 | 0.21 |
ENST00000339023.4 ENST00000354664.4 |
TBC1D3 |
TBC1 domain family, member 3 |
| chrY_+_16636354 | 0.21 |
ENST00000339174.5 |
NLGN4Y |
neuroligin 4, Y-linked |
| chr4_-_39640700 | 0.21 |
ENST00000295958.5 |
SMIM14 |
small integral membrane protein 14 |
| chr17_-_34807272 | 0.21 |
ENST00000535592.1 ENST00000394453.1 |
TBC1D3G |
TBC1 domain family, member 3G |
| chr7_+_29234028 | 0.20 |
ENST00000222792.6 |
CHN2 |
chimerin 2 |
| chr2_+_46926326 | 0.20 |
ENST00000394861.2 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr16_+_19078911 | 0.20 |
ENST00000321998.5 |
COQ7 |
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr1_+_54519242 | 0.19 |
ENST00000234827.1 |
TCEANC2 |
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr14_-_54908043 | 0.17 |
ENST00000556113.1 ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1 |
cornichon family AMPA receptor auxiliary protein 1 |
| chr17_-_34808047 | 0.17 |
ENST00000592614.1 ENST00000591542.1 ENST00000330458.7 ENST00000341264.6 ENST00000592987.1 ENST00000400684.4 |
TBC1D3G TBC1D3H |
TBC1 domain family, member 3G TBC1 domain family, member 3H |
| chr16_+_14802801 | 0.17 |
ENST00000526520.1 ENST00000531598.2 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr1_+_3816936 | 0.17 |
ENST00000413332.1 ENST00000442673.1 ENST00000439488.1 |
RP13-15E13.1 |
long intergenic non-protein coding RNA 1134 |
| chr20_-_52790512 | 0.17 |
ENST00000216862.3 |
CYP24A1 |
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chrX_+_70435044 | 0.17 |
ENST00000374029.1 ENST00000374022.3 ENST00000447581.1 |
GJB1 |
gap junction protein, beta 1, 32kDa |
| chr5_-_179227540 | 0.17 |
ENST00000520875.1 |
MGAT4B |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr10_-_33623564 | 0.17 |
ENST00000374875.1 ENST00000374822.4 |
NRP1 |
neuropilin 1 |
| chr3_+_57541975 | 0.16 |
ENST00000487257.1 ENST00000311180.8 |
PDE12 |
phosphodiesterase 12 |
| chr11_-_65667997 | 0.16 |
ENST00000312562.2 ENST00000534222.1 |
FOSL1 |
FOS-like antigen 1 |
| chr16_+_19079215 | 0.16 |
ENST00000544894.2 ENST00000561858.1 |
COQ7 |
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr16_-_28937027 | 0.16 |
ENST00000358201.4 |
RABEP2 |
rabaptin, RAB GTPase binding effector protein 2 |
| chr7_-_16844611 | 0.16 |
ENST00000401412.1 ENST00000419304.2 |
AGR2 |
anterior gradient 2 |
| chr2_+_113342163 | 0.16 |
ENST00000409719.1 |
CHCHD5 |
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr17_-_34591208 | 0.16 |
ENST00000336331.5 |
TBC1D3C |
TBC1 domain family, member 3C |
| chr17_-_79805146 | 0.16 |
ENST00000415593.1 |
P4HB |
prolyl 4-hydroxylase, beta polypeptide |
| chr7_+_100318423 | 0.16 |
ENST00000252723.2 |
EPO |
erythropoietin |
| chr14_-_20801427 | 0.15 |
ENST00000557665.1 ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1 |
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr12_-_96390063 | 0.15 |
ENST00000541929.1 |
HAL |
histidine ammonia-lyase |
| chr6_+_125475335 | 0.15 |
ENST00000532429.1 ENST00000534199.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr5_-_147211226 | 0.14 |
ENST00000296695.5 |
SPINK1 |
serine peptidase inhibitor, Kazal type 1 |
| chr5_-_147211190 | 0.14 |
ENST00000510027.2 |
SPINK1 |
serine peptidase inhibitor, Kazal type 1 |
| chr11_+_6624970 | 0.14 |
ENST00000420936.2 ENST00000528995.1 |
ILK |
integrin-linked kinase |
| chr1_+_92414952 | 0.14 |
ENST00000449584.1 ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT |
bromodomain, testis-specific |
| chr19_+_56652686 | 0.14 |
ENST00000592949.1 |
ZNF444 |
zinc finger protein 444 |
| chr7_-_151433342 | 0.13 |
ENST00000433631.2 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr19_-_16045665 | 0.13 |
ENST00000248041.8 |
CYP4F11 |
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr19_-_5903714 | 0.13 |
ENST00000586349.1 ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12 NDUFA11 FUT5 |
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chrX_+_149737046 | 0.13 |
ENST00000370396.2 ENST00000542741.1 ENST00000543350.1 ENST00000424519.1 ENST00000413012.2 |
MTM1 |
myotubularin 1 |
| chr7_-_151433393 | 0.13 |
ENST00000492843.1 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr19_-_47987419 | 0.13 |
ENST00000536339.1 ENST00000595554.1 ENST00000600271.1 ENST00000338134.3 |
KPTN |
kaptin (actin binding protein) |
| chr11_-_65667884 | 0.13 |
ENST00000448083.2 ENST00000531493.1 ENST00000532401.1 |
FOSL1 |
FOS-like antigen 1 |
| chr11_-_73882029 | 0.13 |
ENST00000539061.1 |
C2CD3 |
C2 calcium-dependent domain containing 3 |
| chr20_+_55904815 | 0.12 |
ENST00000371263.3 ENST00000345868.4 ENST00000371260.4 ENST00000418127.1 |
SPO11 |
SPO11 meiotic protein covalently bound to DSB |
| chr1_+_100111580 | 0.12 |
ENST00000605497.1 |
PALMD |
palmdelphin |
| chr19_-_16045619 | 0.12 |
ENST00000402119.4 |
CYP4F11 |
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr3_+_184032919 | 0.12 |
ENST00000427845.1 ENST00000342981.4 ENST00000319274.6 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr7_-_105029812 | 0.12 |
ENST00000482897.1 |
SRPK2 |
SRSF protein kinase 2 |
| chr11_+_6624955 | 0.12 |
ENST00000299421.4 ENST00000537806.1 |
ILK |
integrin-linked kinase |
| chr10_-_32345305 | 0.12 |
ENST00000302418.4 |
KIF5B |
kinesin family member 5B |
| chr8_+_119294456 | 0.12 |
ENST00000366457.2 |
AC023590.1 |
Uncharacterized protein |
| chr11_+_6625046 | 0.11 |
ENST00000396751.2 |
ILK |
integrin-linked kinase |
| chr3_+_46618727 | 0.11 |
ENST00000296145.5 |
TDGF1 |
teratocarcinoma-derived growth factor 1 |
| chr13_+_24844819 | 0.11 |
ENST00000399949.2 |
SPATA13 |
spermatogenesis associated 13 |
| chr12_-_63328817 | 0.10 |
ENST00000228705.6 |
PPM1H |
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr19_+_38826415 | 0.10 |
ENST00000410018.1 ENST00000409235.3 |
CATSPERG |
catsper channel auxiliary subunit gamma |
| chr9_+_139839711 | 0.10 |
ENST00000224181.3 |
C8G |
complement component 8, gamma polypeptide |
| chr1_+_200993071 | 0.09 |
ENST00000446333.1 ENST00000458003.1 |
RP11-168O16.1 |
RP11-168O16.1 |
| chr6_-_30524951 | 0.09 |
ENST00000376621.3 |
GNL1 |
guanine nucleotide binding protein-like 1 |
| chr22_-_29663954 | 0.09 |
ENST00000216085.7 |
RHBDD3 |
rhomboid domain containing 3 |
| chr6_-_8064567 | 0.09 |
ENST00000543936.1 ENST00000397457.2 |
BLOC1S5 |
biogenesis of lysosomal organelles complex-1, subunit 5, muted |
| chr1_-_110613276 | 0.09 |
ENST00000369792.4 |
ALX3 |
ALX homeobox 3 |
| chr3_+_132036207 | 0.09 |
ENST00000336375.5 ENST00000495911.1 |
ACPP |
acid phosphatase, prostate |
| chr3_-_122283100 | 0.09 |
ENST00000492382.1 ENST00000462315.1 |
PARP9 |
poly (ADP-ribose) polymerase family, member 9 |
| chr12_+_52695617 | 0.09 |
ENST00000293525.5 |
KRT86 |
keratin 86 |
| chr11_+_28129795 | 0.08 |
ENST00000406787.3 ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15 |
methyltransferase like 15 |
| chr19_-_54693401 | 0.08 |
ENST00000338624.6 |
MBOAT7 |
membrane bound O-acyltransferase domain containing 7 |
| chr2_-_232329186 | 0.08 |
ENST00000322723.4 |
NCL |
nucleolin |
| chr1_-_67519782 | 0.08 |
ENST00000235345.5 |
SLC35D1 |
solute carrier family 35 (UDP-GlcA/UDP-GalNAc transporter), member D1 |
| chr15_-_81282133 | 0.08 |
ENST00000261758.4 |
MESDC2 |
mesoderm development candidate 2 |
| chr4_-_11430221 | 0.08 |
ENST00000514690.1 |
HS3ST1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr3_+_190105909 | 0.07 |
ENST00000456423.1 |
CLDN16 |
claudin 16 |
| chr11_+_64808675 | 0.07 |
ENST00000529996.1 |
SAC3D1 |
SAC3 domain containing 1 |
| chr3_-_178969403 | 0.07 |
ENST00000314235.5 ENST00000392685.2 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr11_+_113779704 | 0.07 |
ENST00000537778.1 |
HTR3B |
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr8_+_12809093 | 0.07 |
ENST00000528753.2 |
KIAA1456 |
KIAA1456 |
| chr12_+_57857475 | 0.07 |
ENST00000528467.1 |
GLI1 |
GLI family zinc finger 1 |
| chr8_+_9046503 | 0.07 |
ENST00000512942.2 |
RP11-10A14.5 |
RP11-10A14.5 |
| chr16_-_28936493 | 0.07 |
ENST00000544477.1 ENST00000357573.6 |
RABEP2 |
rabaptin, RAB GTPase binding effector protein 2 |
| chr16_+_69958887 | 0.07 |
ENST00000568684.1 |
WWP2 |
WW domain containing E3 ubiquitin protein ligase 2 |
| chr5_+_177433973 | 0.07 |
ENST00000507848.1 |
FAM153C |
family with sequence similarity 153, member C |
| chr2_-_170681324 | 0.07 |
ENST00000409340.1 |
METTL5 |
methyltransferase like 5 |
| chr11_-_506316 | 0.07 |
ENST00000532055.1 ENST00000531540.1 |
RNH1 |
ribonuclease/angiogenin inhibitor 1 |
| chr10_-_33624002 | 0.07 |
ENST00000432372.2 |
NRP1 |
neuropilin 1 |
| chr20_+_39765581 | 0.06 |
ENST00000244007.3 |
PLCG1 |
phospholipase C, gamma 1 |
| chr22_+_29664305 | 0.06 |
ENST00000414183.2 ENST00000333395.6 ENST00000455726.1 ENST00000332035.6 |
EWSR1 |
EWS RNA-binding protein 1 |
| chr2_+_102928009 | 0.06 |
ENST00000404917.2 ENST00000447231.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chr11_+_94706804 | 0.06 |
ENST00000335080.5 |
KDM4D |
lysine (K)-specific demethylase 4D |
| chr4_-_36246060 | 0.06 |
ENST00000303965.4 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr3_+_45071622 | 0.06 |
ENST00000428034.1 |
CLEC3B |
C-type lectin domain family 3, member B |
| chr3_+_13521665 | 0.06 |
ENST00000295757.3 ENST00000402259.1 ENST00000402271.1 ENST00000446613.2 ENST00000404548.1 ENST00000404040.1 |
HDAC11 |
histone deacetylase 11 |
| chr12_+_79439405 | 0.06 |
ENST00000552744.1 |
SYT1 |
synaptotagmin I |
| chr5_+_175487692 | 0.06 |
ENST00000510151.1 |
FAM153B |
family with sequence similarity 153, member B |
| chr12_-_96390108 | 0.06 |
ENST00000538703.1 ENST00000261208.3 |
HAL |
histidine ammonia-lyase |
| chr19_+_16830774 | 0.05 |
ENST00000524140.2 |
NWD1 |
NACHT and WD repeat domain containing 1 |
| chr9_-_139343294 | 0.05 |
ENST00000313084.5 |
SEC16A |
SEC16 homolog A (S. cerevisiae) |
| chr19_-_44860820 | 0.05 |
ENST00000354340.4 ENST00000337401.4 ENST00000587909.1 |
ZNF112 |
zinc finger protein 112 |
| chr17_+_43224684 | 0.05 |
ENST00000332499.2 |
HEXIM1 |
hexamethylene bis-acetamide inducible 1 |
| chr4_-_165898768 | 0.05 |
ENST00000329314.5 |
TRIM61 |
tripartite motif containing 61 |
| chr4_-_77997126 | 0.05 |
ENST00000537948.1 ENST00000507788.1 ENST00000237654.4 |
CCNI |
cyclin I |
| chr9_+_139839686 | 0.05 |
ENST00000371634.2 |
C8G |
complement component 8, gamma polypeptide |
| chr15_-_42565606 | 0.05 |
ENST00000307216.6 ENST00000448392.1 |
TMEM87A |
transmembrane protein 87A |
| chr1_+_28099683 | 0.05 |
ENST00000373943.4 |
STX12 |
syntaxin 12 |
| chr19_+_7599597 | 0.05 |
ENST00000414982.3 ENST00000450331.3 |
PNPLA6 |
patatin-like phospholipase domain containing 6 |
| chr5_-_177210399 | 0.05 |
ENST00000510276.1 |
FAM153A |
family with sequence similarity 153, member A |
| chr2_-_85625857 | 0.05 |
ENST00000453973.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr16_+_16043406 | 0.05 |
ENST00000399410.3 ENST00000399408.2 ENST00000346370.5 ENST00000351154.5 ENST00000345148.5 ENST00000349029.5 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
| chr19_-_54804173 | 0.05 |
ENST00000391744.3 ENST00000251390.3 |
LILRA3 |
leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3 |
| chr2_-_73511559 | 0.05 |
ENST00000521871.1 |
FBXO41 |
F-box protein 41 |
| chr20_+_2276639 | 0.05 |
ENST00000381458.5 |
TGM3 |
transglutaminase 3 |
| chr6_+_79577189 | 0.04 |
ENST00000369940.2 |
IRAK1BP1 |
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr11_+_18433840 | 0.04 |
ENST00000541669.1 ENST00000280704.4 |
LDHC |
lactate dehydrogenase C |
| chr19_+_14017116 | 0.04 |
ENST00000589606.1 |
CC2D1A |
coiled-coil and C2 domain containing 1A |
| chr8_+_98900132 | 0.04 |
ENST00000520016.1 |
MATN2 |
matrilin 2 |
| chr16_+_28834303 | 0.04 |
ENST00000340394.8 ENST00000325215.6 ENST00000395547.2 ENST00000336783.4 ENST00000382686.4 ENST00000564304.1 |
ATXN2L |
ataxin 2-like |
| chr16_-_28506840 | 0.04 |
ENST00000569430.1 |
CLN3 |
ceroid-lipofuscinosis, neuronal 3 |
| chr4_-_122854612 | 0.04 |
ENST00000264811.5 |
TRPC3 |
transient receptor potential cation channel, subfamily C, member 3 |
| chr16_+_67207838 | 0.04 |
ENST00000566871.1 ENST00000268605.7 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr12_-_6579833 | 0.04 |
ENST00000396308.3 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr12_+_110011571 | 0.04 |
ENST00000539696.1 ENST00000228510.3 ENST00000392727.3 |
MVK |
mevalonate kinase |
| chr1_+_26348259 | 0.04 |
ENST00000374280.3 |
EXTL1 |
exostosin-like glycosyltransferase 1 |
| chr5_+_175488258 | 0.04 |
ENST00000512862.1 |
FAM153B |
family with sequence similarity 153, member B |
| chr19_+_39936186 | 0.03 |
ENST00000432763.2 ENST00000402194.2 ENST00000601515.1 |
SUPT5H |
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr15_+_41952591 | 0.03 |
ENST00000566718.1 ENST00000219905.7 ENST00000389936.4 ENST00000545763.1 |
MGA |
MGA, MAX dimerization protein |
| chr7_-_36764142 | 0.03 |
ENST00000258749.5 ENST00000535891.1 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
| chr1_+_27561104 | 0.03 |
ENST00000361771.3 |
WDTC1 |
WD and tetratricopeptide repeats 1 |
| chr16_+_640201 | 0.03 |
ENST00000563109.1 |
RAB40C |
RAB40C, member RAS oncogene family |
| chr12_-_114404111 | 0.03 |
ENST00000545145.2 ENST00000392561.3 ENST00000261741.5 |
RBM19 |
RNA binding motif protein 19 |
| chr16_+_67207872 | 0.03 |
ENST00000563258.1 ENST00000568146.1 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr9_+_111624577 | 0.03 |
ENST00000333999.3 |
ACTL7A |
actin-like 7A |
| chr10_+_119301928 | 0.03 |
ENST00000553456.3 |
EMX2 |
empty spiracles homeobox 2 |
| chr15_+_69307028 | 0.03 |
ENST00000388866.3 ENST00000530406.2 |
NOX5 |
NADPH oxidase, EF-hand calcium binding domain 5 |
| chr19_-_13261160 | 0.03 |
ENST00000343587.5 ENST00000591197.1 |
STX10 |
syntaxin 10 |
| chr1_-_19426149 | 0.03 |
ENST00000429347.2 |
UBR4 |
ubiquitin protein ligase E3 component n-recognin 4 |
| chr5_-_177209832 | 0.03 |
ENST00000393518.3 ENST00000505531.1 ENST00000503567.1 |
FAM153A |
family with sequence similarity 153, member A |
| chr3_-_3151664 | 0.03 |
ENST00000256452.3 ENST00000311981.8 ENST00000430514.2 ENST00000456302.1 |
IL5RA |
interleukin 5 receptor, alpha |
| chr11_+_124609823 | 0.03 |
ENST00000412681.2 |
NRGN |
neurogranin (protein kinase C substrate, RC3) |
| chr22_+_29664248 | 0.03 |
ENST00000406548.1 ENST00000437155.2 ENST00000415761.1 ENST00000331029.7 |
EWSR1 |
EWS RNA-binding protein 1 |
| chr11_+_63998198 | 0.03 |
ENST00000321460.5 |
DNAJC4 |
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr8_-_93115445 | 0.02 |
ENST00000523629.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr3_+_93781728 | 0.02 |
ENST00000314622.4 |
NSUN3 |
NOP2/Sun domain family, member 3 |
| chr12_-_6579808 | 0.02 |
ENST00000535180.1 ENST00000400911.3 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr6_-_32145101 | 0.02 |
ENST00000375104.2 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr14_+_94640633 | 0.02 |
ENST00000304338.3 |
PPP4R4 |
protein phosphatase 4, regulatory subunit 4 |
| chr14_+_22963806 | 0.02 |
ENST00000390493.1 |
TRAJ44 |
T cell receptor alpha joining 44 |
| chr6_+_24775153 | 0.02 |
ENST00000356509.3 ENST00000230056.3 |
GMNN |
geminin, DNA replication inhibitor |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.7 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.3 | 0.8 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 0.8 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.4 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.7 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.1 | 0.3 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.1 | 0.8 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.3 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 1.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 0.5 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.0 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.6 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 1.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0097180 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.2 | 1.1 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.1 | 0.5 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.3 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.8 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) sorting endosome(GO:0097443) |
| 0.0 | 0.3 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) negative regulation of cell fate specification(GO:0009996) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 0.7 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 0.4 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.5 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.1 | 0.8 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.1 | 1.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.5 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 0.3 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.3 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 0.8 | GO:0051918 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 0.7 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.1 | 0.3 | GO:0042361 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.1 | 0.4 | GO:0070417 | cellular response to cold(GO:0070417) nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 0.3 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.1 | 0.2 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.1 | 0.2 | GO:0001207 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 0.5 | GO:0051581 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.7 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.8 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 0.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.7 | GO:0097296 | activation of NF-kappaB-inducing kinase activity(GO:0007250) activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.1 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.6 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.2 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0035617 | cytoplasm organization(GO:0007028) stress granule disassembly(GO:0035617) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.2 | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway(GO:0035791) |
| 0.0 | 1.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0021938 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0015868 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.0 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.0 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.4 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0090220 | meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.3 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.0 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 1.8 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.0 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 0.0 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |