Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
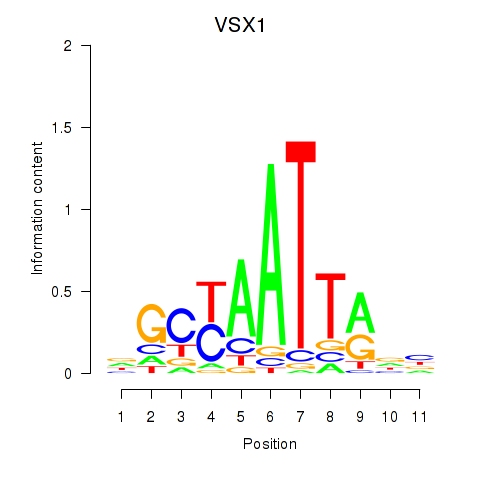
Results for VSX1
Z-value: 0.57
Transcription factors associated with VSX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VSX1
|
ENSG00000100987.10 | VSX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VSX1 | hg19_v2_chr20_-_25062767_25062779 | -0.31 | 2.4e-01 | Click! |
Activity profile of VSX1 motif
Sorted Z-values of VSX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of VSX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_31783291 | 1.20 |
ENST00000375651.5 ENST00000608703.1 ENST00000458062.2 |
HSPA1A |
heat shock 70kDa protein 1A |
| chr13_-_110438914 | 0.73 |
ENST00000375856.3 |
IRS2 |
insulin receptor substrate 2 |
| chr11_-_62521614 | 0.70 |
ENST00000527994.1 ENST00000394807.3 |
ZBTB3 |
zinc finger and BTB domain containing 3 |
| chr6_+_26124373 | 0.66 |
ENST00000377791.2 ENST00000602637.1 |
HIST1H2AC |
histone cluster 1, H2ac |
| chr6_-_32145861 | 0.63 |
ENST00000336984.6 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr10_-_126107482 | 0.62 |
ENST00000368845.5 ENST00000539214.1 |
OAT |
ornithine aminotransferase |
| chr19_+_45394477 | 0.61 |
ENST00000252487.5 ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40 |
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr6_+_26199737 | 0.60 |
ENST00000359985.1 |
HIST1H2BF |
histone cluster 1, H2bf |
| chr6_-_26199499 | 0.60 |
ENST00000377831.5 |
HIST1H3D |
histone cluster 1, H3d |
| chr15_+_89631647 | 0.58 |
ENST00000569550.1 ENST00000565066.1 ENST00000565973.1 |
ABHD2 |
abhydrolase domain containing 2 |
| chr10_-_5046042 | 0.58 |
ENST00000421196.3 ENST00000455190.1 |
AKR1C2 |
aldo-keto reductase family 1, member C2 |
| chr19_+_39421556 | 0.57 |
ENST00000407800.2 ENST00000402029.3 |
MRPS12 |
mitochondrial ribosomal protein S12 |
| chr7_+_12727250 | 0.57 |
ENST00000404894.1 |
ARL4A |
ADP-ribosylation factor-like 4A |
| chr6_-_26124138 | 0.56 |
ENST00000314332.5 ENST00000396984.1 |
HIST1H2BC |
histone cluster 1, H2bc |
| chr2_-_192711968 | 0.55 |
ENST00000304141.4 |
SDPR |
serum deprivation response |
| chr6_-_26199471 | 0.51 |
ENST00000341023.1 |
HIST1H2AD |
histone cluster 1, H2ad |
| chr17_-_79980734 | 0.51 |
ENST00000584600.1 ENST00000584347.1 ENST00000580435.1 ENST00000306704.6 ENST00000392359.3 |
STRA13 |
stimulated by retinoic acid 13 |
| chr11_-_33913708 | 0.48 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr1_-_167905225 | 0.45 |
ENST00000367846.4 |
MPC2 |
mitochondrial pyruvate carrier 2 |
| chr12_+_81110684 | 0.44 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr21_-_34914394 | 0.44 |
ENST00000361093.5 ENST00000381815.4 |
GART |
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr19_-_15529790 | 0.43 |
ENST00000596195.1 ENST00000595067.1 ENST00000595465.2 ENST00000397410.5 ENST00000600247.1 |
AKAP8L |
A kinase (PRKA) anchor protein 8-like |
| chr5_+_53751445 | 0.43 |
ENST00000302005.1 |
HSPB3 |
heat shock 27kDa protein 3 |
| chr11_-_32457176 | 0.43 |
ENST00000332351.3 |
WT1 |
Wilms tumor 1 |
| chr12_-_49075941 | 0.42 |
ENST00000553086.1 ENST00000548304.1 |
KANSL2 |
KAT8 regulatory NSL complex subunit 2 |
| chr2_+_234637754 | 0.42 |
ENST00000482026.1 ENST00000609767.1 |
UGT1A3 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr3_+_118905564 | 0.42 |
ENST00000460625.1 |
UPK1B |
uroplakin 1B |
| chr6_+_31795506 | 0.42 |
ENST00000375650.3 |
HSPA1B |
heat shock 70kDa protein 1B |
| chr11_-_122932730 | 0.41 |
ENST00000532182.1 ENST00000524590.1 ENST00000528292.1 ENST00000533540.1 ENST00000525463.1 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr11_+_57480046 | 0.41 |
ENST00000378312.4 ENST00000278422.4 |
TMX2 |
thioredoxin-related transmembrane protein 2 |
| chr4_+_1283639 | 0.40 |
ENST00000303400.4 ENST00000505177.2 ENST00000503653.1 ENST00000264750.6 ENST00000502558.1 ENST00000452175.2 ENST00000514708.1 |
MAEA |
macrophage erythroblast attacher |
| chr10_-_99447024 | 0.39 |
ENST00000370626.3 |
AVPI1 |
arginine vasopressin-induced 1 |
| chr15_+_89631381 | 0.39 |
ENST00000352732.5 |
ABHD2 |
abhydrolase domain containing 2 |
| chr4_-_143226979 | 0.39 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr4_-_143227088 | 0.39 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr19_+_9945962 | 0.38 |
ENST00000587625.1 ENST00000247970.4 ENST00000588695.1 |
PIN1 |
peptidylprolyl cis/trans isomerase, NIMA-interacting 1 |
| chr5_+_44809027 | 0.36 |
ENST00000507110.1 |
MRPS30 |
mitochondrial ribosomal protein S30 |
| chr1_+_167905894 | 0.36 |
ENST00000367843.3 ENST00000432587.2 ENST00000312263.6 |
DCAF6 |
DDB1 and CUL4 associated factor 6 |
| chr11_-_3818688 | 0.36 |
ENST00000355260.3 ENST00000397004.4 ENST00000397007.4 ENST00000532475.1 |
NUP98 |
nucleoporin 98kDa |
| chr7_-_99699538 | 0.36 |
ENST00000343023.6 ENST00000303887.5 |
MCM7 |
minichromosome maintenance complex component 7 |
| chr6_-_27860956 | 0.36 |
ENST00000359611.2 |
HIST1H2AM |
histone cluster 1, H2am |
| chr11_-_122933043 | 0.35 |
ENST00000534624.1 ENST00000453788.2 ENST00000527387.1 |
HSPA8 |
heat shock 70kDa protein 8 |
| chr11_-_59383617 | 0.34 |
ENST00000263847.1 |
OSBP |
oxysterol binding protein |
| chr12_+_110011571 | 0.34 |
ENST00000539696.1 ENST00000228510.3 ENST00000392727.3 |
MVK |
mevalonate kinase |
| chr13_-_19755975 | 0.33 |
ENST00000400113.3 |
TUBA3C |
tubulin, alpha 3c |
| chr3_+_47324424 | 0.32 |
ENST00000437353.1 ENST00000232766.5 ENST00000455924.2 |
KLHL18 |
kelch-like family member 18 |
| chr17_-_42277203 | 0.32 |
ENST00000587097.1 |
ATXN7L3 |
ataxin 7-like 3 |
| chr3_+_152552685 | 0.32 |
ENST00000305097.3 |
P2RY1 |
purinergic receptor P2Y, G-protein coupled, 1 |
| chr14_+_104182061 | 0.32 |
ENST00000216602.6 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr14_+_29236269 | 0.31 |
ENST00000313071.4 |
FOXG1 |
forkhead box G1 |
| chr14_+_104182105 | 0.31 |
ENST00000311141.2 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr1_+_154947126 | 0.30 |
ENST00000368439.1 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
| chr7_-_44613494 | 0.29 |
ENST00000431640.1 ENST00000258772.5 |
DDX56 |
DEAD (Asp-Glu-Ala-Asp) box helicase 56 |
| chr11_-_2906979 | 0.29 |
ENST00000380725.1 ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C |
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr11_+_67798363 | 0.28 |
ENST00000525628.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr10_+_5005598 | 0.28 |
ENST00000442997.1 |
AKR1C1 |
aldo-keto reductase family 1, member C1 |
| chr12_-_49076002 | 0.28 |
ENST00000357861.3 ENST00000550347.1 ENST00000420613.2 ENST00000550931.1 ENST00000550870.1 |
KANSL2 |
KAT8 regulatory NSL complex subunit 2 |
| chr19_+_17420340 | 0.28 |
ENST00000359866.4 |
DDA1 |
DET1 and DDB1 associated 1 |
| chr2_-_24270217 | 0.27 |
ENST00000295148.4 ENST00000406895.3 |
C2orf44 |
chromosome 2 open reading frame 44 |
| chr1_+_154947148 | 0.27 |
ENST00000368436.1 ENST00000308987.5 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
| chr4_+_147096837 | 0.27 |
ENST00000296581.5 ENST00000502781.1 |
LSM6 |
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr15_+_41523417 | 0.26 |
ENST00000560397.1 |
CHP1 |
calcineurin-like EF-hand protein 1 |
| chr11_-_66056478 | 0.26 |
ENST00000431556.2 ENST00000528575.1 |
YIF1A |
Yip1 interacting factor homolog A (S. cerevisiae) |
| chrX_-_47509887 | 0.26 |
ENST00000247161.3 ENST00000592066.1 ENST00000376983.3 |
ELK1 |
ELK1, member of ETS oncogene family |
| chr12_+_28410128 | 0.26 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr19_+_19303720 | 0.26 |
ENST00000392324.4 |
RFXANK |
regulatory factor X-associated ankyrin-containing protein |
| chr5_-_82969405 | 0.25 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chr11_+_67798090 | 0.24 |
ENST00000313468.5 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chrX_+_19373700 | 0.24 |
ENST00000379804.1 |
PDHA1 |
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr19_-_13227534 | 0.24 |
ENST00000588229.1 ENST00000357720.4 |
TRMT1 |
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr14_+_58711539 | 0.24 |
ENST00000216455.4 ENST00000412908.2 ENST00000557508.1 |
PSMA3 |
proteasome (prosome, macropain) subunit, alpha type, 3 |
| chr19_+_19303572 | 0.24 |
ENST00000407360.3 ENST00000540981.1 |
RFXANK |
regulatory factor X-associated ankyrin-containing protein |
| chrX_+_151999511 | 0.24 |
ENST00000370274.3 ENST00000440023.1 ENST00000432467.1 |
NSDHL |
NAD(P) dependent steroid dehydrogenase-like |
| chr5_-_150138061 | 0.23 |
ENST00000521533.1 ENST00000424236.1 |
DCTN4 |
dynactin 4 (p62) |
| chr10_-_127505167 | 0.23 |
ENST00000368786.1 |
UROS |
uroporphyrinogen III synthase |
| chr7_+_6048856 | 0.23 |
ENST00000223029.3 ENST00000400479.2 ENST00000395236.2 |
AIMP2 |
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr14_+_32798547 | 0.23 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr11_-_3818932 | 0.22 |
ENST00000324932.7 ENST00000359171.4 |
NUP98 |
nucleoporin 98kDa |
| chr12_+_106751436 | 0.22 |
ENST00000228347.4 |
POLR3B |
polymerase (RNA) III (DNA directed) polypeptide B |
| chr19_-_13227463 | 0.22 |
ENST00000437766.1 ENST00000221504.8 |
TRMT1 |
tRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr15_+_43803143 | 0.22 |
ENST00000382031.1 |
MAP1A |
microtubule-associated protein 1A |
| chr3_-_186288097 | 0.21 |
ENST00000446782.1 |
TBCCD1 |
TBCC domain containing 1 |
| chr19_-_39421377 | 0.21 |
ENST00000430193.3 ENST00000600042.1 ENST00000221431.6 |
SARS2 |
seryl-tRNA synthetase 2, mitochondrial |
| chr1_+_24286287 | 0.21 |
ENST00000334351.7 ENST00000374468.1 |
PNRC2 |
proline-rich nuclear receptor coactivator 2 |
| chr7_+_55433131 | 0.20 |
ENST00000254770.2 |
LANCL2 |
LanC lantibiotic synthetase component C-like 2 (bacterial) |
| chr10_-_105845674 | 0.20 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr1_-_169455169 | 0.20 |
ENST00000367804.4 ENST00000236137.5 |
SLC19A2 |
solute carrier family 19 (thiamine transporter), member 2 |
| chr11_+_67798114 | 0.19 |
ENST00000453471.2 ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr10_+_1095416 | 0.19 |
ENST00000358220.1 |
WDR37 |
WD repeat domain 37 |
| chr9_-_99801592 | 0.19 |
ENST00000259470.5 |
CTSV |
cathepsin V |
| chr1_-_35658736 | 0.19 |
ENST00000357214.5 |
SFPQ |
splicing factor proline/glutamine-rich |
| chr2_-_179672142 | 0.19 |
ENST00000342992.6 ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN |
titin |
| chr6_+_30687978 | 0.19 |
ENST00000327892.8 ENST00000435534.1 |
TUBB |
tubulin, beta class I |
| chr6_+_160211481 | 0.18 |
ENST00000367034.4 |
MRPL18 |
mitochondrial ribosomal protein L18 |
| chr1_-_11322551 | 0.18 |
ENST00000361445.4 |
MTOR |
mechanistic target of rapamycin (serine/threonine kinase) |
| chr17_+_46970134 | 0.18 |
ENST00000503641.1 ENST00000514808.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr7_-_44887620 | 0.18 |
ENST00000349299.3 ENST00000521529.1 ENST00000308153.4 ENST00000350771.3 ENST00000222690.6 ENST00000381124.5 ENST00000437072.1 ENST00000446531.1 |
H2AFV |
H2A histone family, member V |
| chr6_-_31782813 | 0.17 |
ENST00000375654.4 |
HSPA1L |
heat shock 70kDa protein 1-like |
| chr11_+_560956 | 0.17 |
ENST00000397582.3 ENST00000344375.4 ENST00000397583.3 |
RASSF7 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr18_-_33702078 | 0.17 |
ENST00000586829.1 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr16_-_31085514 | 0.17 |
ENST00000300849.4 |
ZNF668 |
zinc finger protein 668 |
| chr6_+_27861190 | 0.17 |
ENST00000303806.4 |
HIST1H2BO |
histone cluster 1, H2bo |
| chr17_-_47785504 | 0.16 |
ENST00000514907.1 ENST00000503334.1 ENST00000508520.1 |
SLC35B1 |
solute carrier family 35, member B1 |
| chr15_-_43785303 | 0.15 |
ENST00000382039.3 ENST00000450115.2 ENST00000382044.4 |
TP53BP1 |
tumor protein p53 binding protein 1 |
| chr17_+_46970178 | 0.15 |
ENST00000393366.2 ENST00000506855.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr16_+_68119440 | 0.15 |
ENST00000346183.3 ENST00000329524.4 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr17_+_46970127 | 0.15 |
ENST00000355938.5 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chrX_+_154444643 | 0.15 |
ENST00000286428.5 |
VBP1 |
von Hippel-Lindau binding protein 1 |
| chr16_+_68119247 | 0.15 |
ENST00000575270.1 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr5_+_151151471 | 0.15 |
ENST00000394123.3 ENST00000543466.1 |
G3BP1 |
GTPase activating protein (SH3 domain) binding protein 1 |
| chr16_+_23652773 | 0.14 |
ENST00000563998.1 ENST00000568589.1 ENST00000568272.1 |
DCTN5 |
dynactin 5 (p25) |
| chrX_-_151999269 | 0.14 |
ENST00000370277.3 |
CETN2 |
centrin, EF-hand protein, 2 |
| chr11_+_3819049 | 0.14 |
ENST00000396986.2 ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2 |
post-GPI attachment to proteins 2 |
| chr14_-_104181771 | 0.14 |
ENST00000554913.1 ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3 |
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr8_+_105235572 | 0.14 |
ENST00000523362.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr2_-_217560248 | 0.14 |
ENST00000233813.4 |
IGFBP5 |
insulin-like growth factor binding protein 5 |
| chr10_+_85899196 | 0.13 |
ENST00000372134.3 |
GHITM |
growth hormone inducible transmembrane protein |
| chr19_+_54641444 | 0.13 |
ENST00000221232.5 ENST00000358389.3 |
CNOT3 |
CCR4-NOT transcription complex, subunit 3 |
| chr3_+_179280668 | 0.13 |
ENST00000429709.2 ENST00000450518.2 ENST00000392662.1 ENST00000490364.1 |
ACTL6A |
actin-like 6A |
| chr1_-_154946825 | 0.13 |
ENST00000368453.4 ENST00000368450.1 ENST00000366442.2 |
SHC1 |
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr1_-_67266939 | 0.13 |
ENST00000304526.2 |
INSL5 |
insulin-like 5 |
| chr17_-_47785265 | 0.13 |
ENST00000511763.1 ENST00000515850.1 ENST00000415270.2 ENST00000240333.6 |
SLC35B1 |
solute carrier family 35, member B1 |
| chr2_+_10183651 | 0.13 |
ENST00000305883.1 |
KLF11 |
Kruppel-like factor 11 |
| chr1_-_149900122 | 0.13 |
ENST00000271628.8 |
SF3B4 |
splicing factor 3b, subunit 4, 49kDa |
| chr9_+_140135665 | 0.13 |
ENST00000340384.4 |
TUBB4B |
tubulin, beta 4B class IVb |
| chr19_-_15236173 | 0.13 |
ENST00000527093.1 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
| chr19_+_54619125 | 0.13 |
ENST00000445811.1 ENST00000419967.1 ENST00000445124.1 ENST00000447810.1 |
PRPF31 |
pre-mRNA processing factor 31 |
| chr14_-_78083112 | 0.12 |
ENST00000216484.2 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
| chr15_+_41523335 | 0.12 |
ENST00000334660.5 |
CHP1 |
calcineurin-like EF-hand protein 1 |
| chr17_-_74733404 | 0.12 |
ENST00000508921.3 ENST00000583836.1 ENST00000358156.6 ENST00000392485.2 ENST00000359995.5 |
SRSF2 |
serine/arginine-rich splicing factor 2 |
| chr5_+_159436120 | 0.12 |
ENST00000522793.1 ENST00000231238.5 |
TTC1 |
tetratricopeptide repeat domain 1 |
| chr1_+_151138526 | 0.12 |
ENST00000368902.1 |
SCNM1 |
sodium channel modifier 1 |
| chr7_-_25019760 | 0.12 |
ENST00000352860.1 ENST00000353930.1 ENST00000431825.2 ENST00000313367.2 |
OSBPL3 |
oxysterol binding protein-like 3 |
| chr4_-_39979576 | 0.12 |
ENST00000303538.8 ENST00000503396.1 |
PDS5A |
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr17_-_40829026 | 0.12 |
ENST00000412503.1 |
PLEKHH3 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr19_+_34287751 | 0.12 |
ENST00000590771.1 ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
| chr1_+_44115814 | 0.12 |
ENST00000372396.3 |
KDM4A |
lysine (K)-specific demethylase 4A |
| chr19_+_11649532 | 0.12 |
ENST00000252456.2 ENST00000592923.1 ENST00000535659.2 |
CNN1 |
calponin 1, basic, smooth muscle |
| chr19_+_13001840 | 0.11 |
ENST00000222214.5 ENST00000589039.1 ENST00000591470.1 ENST00000457854.1 ENST00000422947.2 ENST00000588905.1 ENST00000587072.1 |
GCDH |
glutaryl-CoA dehydrogenase |
| chr15_-_77363513 | 0.11 |
ENST00000267970.4 |
TSPAN3 |
tetraspanin 3 |
| chr16_+_69345243 | 0.11 |
ENST00000254950.11 |
VPS4A |
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chrM_+_4431 | 0.11 |
ENST00000361453.3 |
MT-ND2 |
mitochondrially encoded NADH dehydrogenase 2 |
| chr15_-_77363441 | 0.11 |
ENST00000346495.2 ENST00000424443.3 ENST00000561277.1 |
TSPAN3 |
tetraspanin 3 |
| chr15_-_43802769 | 0.11 |
ENST00000263801.3 |
TP53BP1 |
tumor protein p53 binding protein 1 |
| chr8_+_9413410 | 0.11 |
ENST00000520408.1 ENST00000310430.6 ENST00000522110.1 |
TNKS |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr6_+_32146131 | 0.11 |
ENST00000375094.3 |
RNF5 |
ring finger protein 5, E3 ubiquitin protein ligase |
| chr2_-_55276320 | 0.11 |
ENST00000357376.3 |
RTN4 |
reticulon 4 |
| chr2_-_153574480 | 0.11 |
ENST00000410080.1 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr6_-_94129244 | 0.11 |
ENST00000369303.4 ENST00000369297.1 |
EPHA7 |
EPH receptor A7 |
| chr5_+_177631523 | 0.11 |
ENST00000506339.1 ENST00000355836.5 ENST00000514633.1 ENST00000515193.1 ENST00000506259.1 ENST00000504898.1 |
HNRNPAB |
heterogeneous nuclear ribonucleoprotein A/B |
| chr2_-_201828356 | 0.10 |
ENST00000234296.2 |
ORC2 |
origin recognition complex, subunit 2 |
| chrX_-_153363188 | 0.10 |
ENST00000303391.6 |
MECP2 |
methyl CpG binding protein 2 (Rett syndrome) |
| chr7_-_6048650 | 0.10 |
ENST00000382321.4 ENST00000406569.3 |
PMS2 |
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr10_-_1095050 | 0.10 |
ENST00000381344.3 |
IDI1 |
isopentenyl-diphosphate delta isomerase 1 |
| chr3_+_149191723 | 0.10 |
ENST00000305354.4 |
TM4SF4 |
transmembrane 4 L six family member 4 |
| chr7_+_74072288 | 0.10 |
ENST00000443166.1 |
GTF2I |
general transcription factor IIi |
| chr11_-_4629388 | 0.10 |
ENST00000526337.1 ENST00000300747.5 |
TRIM68 |
tripartite motif containing 68 |
| chr16_-_11036300 | 0.10 |
ENST00000331808.4 |
DEXI |
Dexi homolog (mouse) |
| chr8_-_145159083 | 0.10 |
ENST00000398712.2 |
SHARPIN |
SHANK-associated RH domain interactor |
| chr1_-_190446759 | 0.10 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr17_+_45727204 | 0.10 |
ENST00000290158.4 |
KPNB1 |
karyopherin (importin) beta 1 |
| chr16_+_68119324 | 0.10 |
ENST00000349223.5 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr22_-_42486747 | 0.10 |
ENST00000602404.1 |
NDUFA6 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa |
| chr2_+_65454926 | 0.10 |
ENST00000542850.1 ENST00000377982.4 |
ACTR2 |
ARP2 actin-related protein 2 homolog (yeast) |
| chr17_+_79650962 | 0.10 |
ENST00000329138.4 |
HGS |
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr3_-_33481835 | 0.09 |
ENST00000283629.3 |
UBP1 |
upstream binding protein 1 (LBP-1a) |
| chr3_-_38992052 | 0.09 |
ENST00000302328.3 ENST00000450244.1 ENST00000444237.2 |
SCN11A |
sodium channel, voltage-gated, type XI, alpha subunit |
| chr15_-_77363375 | 0.09 |
ENST00000559494.1 |
TSPAN3 |
tetraspanin 3 |
| chr5_+_177631497 | 0.09 |
ENST00000358344.3 |
HNRNPAB |
heterogeneous nuclear ribonucleoprotein A/B |
| chr4_+_87515454 | 0.09 |
ENST00000427191.2 ENST00000436978.1 ENST00000502971.1 |
PTPN13 |
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) |
| chr20_+_49126881 | 0.09 |
ENST00000371621.3 ENST00000541713.1 |
PTPN1 |
protein tyrosine phosphatase, non-receptor type 1 |
| chr19_-_14606900 | 0.09 |
ENST00000393029.3 ENST00000393028.1 ENST00000393033.4 ENST00000345425.2 ENST00000586027.1 ENST00000591349.1 ENST00000587210.1 |
GIPC1 |
GIPC PDZ domain containing family, member 1 |
| chr1_-_47655686 | 0.09 |
ENST00000294338.2 |
PDZK1IP1 |
PDZK1 interacting protein 1 |
| chr21_+_35014706 | 0.08 |
ENST00000399353.1 ENST00000444491.1 ENST00000381318.3 |
ITSN1 |
intersectin 1 (SH3 domain protein) |
| chr10_+_74451883 | 0.08 |
ENST00000373053.3 ENST00000357157.6 |
MCU |
mitochondrial calcium uniporter |
| chr7_-_99716952 | 0.08 |
ENST00000523306.1 ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr6_-_26271612 | 0.08 |
ENST00000305910.3 |
HIST1H3G |
histone cluster 1, H3g |
| chr14_+_32798462 | 0.08 |
ENST00000280979.4 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr6_-_33385902 | 0.08 |
ENST00000374500.5 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr2_-_70780770 | 0.08 |
ENST00000444975.1 ENST00000445399.1 ENST00000418333.2 |
TGFA |
transforming growth factor, alpha |
| chr1_-_23670817 | 0.08 |
ENST00000478691.1 |
HNRNPR |
heterogeneous nuclear ribonucleoprotein R |
| chr21_-_42219065 | 0.08 |
ENST00000400454.1 |
DSCAM |
Down syndrome cell adhesion molecule |
| chrY_+_23698778 | 0.08 |
ENST00000303902.5 |
RBMY1A1 |
RNA binding motif protein, Y-linked, family 1, member A1 |
| chr7_-_72971934 | 0.07 |
ENST00000411832.1 |
BCL7B |
B-cell CLL/lymphoma 7B |
| chr6_-_33385870 | 0.07 |
ENST00000488034.1 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr1_-_6052463 | 0.07 |
ENST00000378156.4 |
NPHP4 |
nephronophthisis 4 |
| chr12_+_98909351 | 0.07 |
ENST00000343315.5 ENST00000266732.4 ENST00000393053.2 |
TMPO |
thymopoietin |
| chr6_-_32157947 | 0.07 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr11_+_64018955 | 0.07 |
ENST00000279230.6 ENST00000540288.1 ENST00000325234.5 |
PLCB3 |
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr17_-_40828969 | 0.07 |
ENST00000591022.1 ENST00000587627.1 ENST00000293349.6 |
PLEKHH3 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr5_+_33440802 | 0.06 |
ENST00000502553.1 ENST00000514259.1 ENST00000265112.3 |
TARS |
threonyl-tRNA synthetase |
| chr10_+_70661014 | 0.06 |
ENST00000373585.3 |
DDX50 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr12_-_14849470 | 0.06 |
ENST00000261170.3 |
GUCY2C |
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chrX_+_70503037 | 0.06 |
ENST00000535149.1 |
NONO |
non-POU domain containing, octamer-binding |
| chr3_-_44519131 | 0.06 |
ENST00000425708.2 ENST00000396077.2 |
ZNF445 |
zinc finger protein 445 |
| chr4_-_168155169 | 0.06 |
ENST00000534949.1 ENST00000535728.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr10_+_98064085 | 0.06 |
ENST00000419175.1 ENST00000371174.2 |
DNTT |
DNA nucleotidylexotransferase |
| chr6_-_111927449 | 0.06 |
ENST00000368761.5 ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr17_+_1733276 | 0.06 |
ENST00000254719.5 |
RPA1 |
replication protein A1, 70kDa |
| chr6_-_26032288 | 0.06 |
ENST00000244661.2 |
HIST1H3B |
histone cluster 1, H3b |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.9 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.8 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 0.6 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.0 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.3 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 2.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 1.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.4 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.9 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.2 | 0.8 | GO:1904764 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.1 | 0.9 | GO:1903960 | negative regulation of anion transmembrane transport(GO:1903960) |
| 0.1 | 0.9 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 0.6 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.5 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.4 | GO:0072166 | posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.1 | 0.6 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.1 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 0.3 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 0.4 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.1 | 0.4 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.1 | 0.3 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.2 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 | 0.2 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.1 | 0.2 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.2 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.1 | 0.2 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.1 | 0.3 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.1 | 0.1 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.1 | 0.4 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.4 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.4 | GO:0070885 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.7 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.3 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.0 | 0.2 | GO:1990834 | response to odorant(GO:1990834) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 1.0 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.4 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.1 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.0 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0010038 | response to metal ion(GO:0010038) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.5 | GO:1902894 | negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.0 | 0.3 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 1.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.3 | GO:0044144 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.0 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.6 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.9 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:2000172 | regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.1 | GO:1900045 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0006568 | lysine catabolic process(GO:0006554) tryptophan metabolic process(GO:0006568) |
| 0.0 | 0.2 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.0 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.8 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.9 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.6 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.2 | 0.8 | GO:0034597 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.9 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.1 | 0.5 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.6 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.1 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.4 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 0.4 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.3 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 1.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.2 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.2 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.2 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.1 | 0.2 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.1 | 0.7 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 0.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.3 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.6 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |