Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
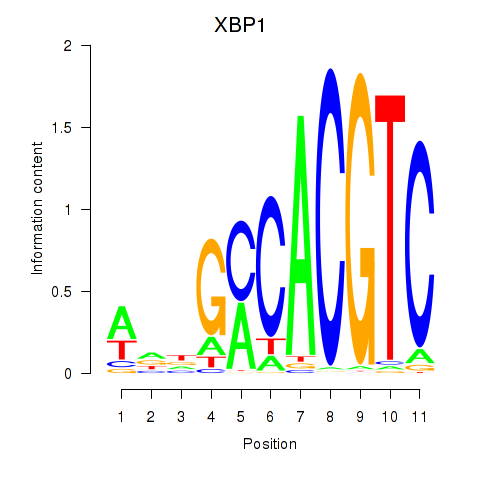
Results for XBP1
Z-value: 2.21
Transcription factors associated with XBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
XBP1
|
ENSG00000100219.12 | XBP1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| XBP1 | hg19_v2_chr22_-_29196030_29196121 | -0.29 | 2.8e-01 | Click! |
Activity profile of XBP1 motif
Sorted Z-values of XBP1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of XBP1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_94023873 | 21.01 |
ENST00000297268.6 |
COL1A2 |
collagen, type I, alpha 2 |
| chr10_+_89419370 | 7.45 |
ENST00000361175.4 ENST00000456849.1 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr5_-_121413974 | 6.26 |
ENST00000231004.4 |
LOX |
lysyl oxidase |
| chr22_+_38864041 | 5.84 |
ENST00000216014.4 ENST00000409006.3 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr12_-_106641728 | 5.72 |
ENST00000378026.4 |
CKAP4 |
cytoskeleton-associated protein 4 |
| chr11_+_69455855 | 5.44 |
ENST00000227507.2 ENST00000536559.1 |
CCND1 |
cyclin D1 |
| chr5_+_119799927 | 3.92 |
ENST00000407149.2 ENST00000379551.2 |
PRR16 |
proline rich 16 |
| chr15_-_60690932 | 3.85 |
ENST00000559818.1 |
ANXA2 |
annexin A2 |
| chr3_-_149688502 | 3.84 |
ENST00000481767.1 ENST00000475518.1 |
PFN2 |
profilin 2 |
| chr3_-_149688896 | 3.81 |
ENST00000239940.7 |
PFN2 |
profilin 2 |
| chr7_+_128379346 | 3.78 |
ENST00000535011.2 ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU |
calumenin |
| chr3_-_57583185 | 3.57 |
ENST00000463880.1 |
ARF4 |
ADP-ribosylation factor 4 |
| chr7_-_19157248 | 3.34 |
ENST00000242261.5 |
TWIST1 |
twist family bHLH transcription factor 1 |
| chr7_+_128379449 | 3.30 |
ENST00000479257.1 |
CALU |
calumenin |
| chr7_-_27183263 | 3.26 |
ENST00000222726.3 |
HOXA5 |
homeobox A5 |
| chr3_-_57583052 | 3.19 |
ENST00000496292.1 ENST00000489843.1 |
ARF4 |
ADP-ribosylation factor 4 |
| chr3_+_105086056 | 3.14 |
ENST00000472644.2 |
ALCAM |
activated leukocyte cell adhesion molecule |
| chr3_-_57583130 | 3.03 |
ENST00000303436.6 |
ARF4 |
ADP-ribosylation factor 4 |
| chr12_-_56123444 | 2.73 |
ENST00000546457.1 ENST00000549117.1 |
CD63 |
CD63 molecule |
| chr1_-_144932014 | 2.68 |
ENST00000529945.1 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr20_-_17662878 | 2.58 |
ENST00000377813.1 ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1 |
ribosome binding protein 1 |
| chr5_-_9546180 | 2.54 |
ENST00000382496.5 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr3_+_171758344 | 2.51 |
ENST00000336824.4 ENST00000423424.1 |
FNDC3B |
fibronectin type III domain containing 3B |
| chr14_+_100259666 | 2.40 |
ENST00000262233.6 ENST00000334192.4 |
EML1 |
echinoderm microtubule associated protein like 1 |
| chr6_+_116692102 | 2.39 |
ENST00000359564.2 |
DSE |
dermatan sulfate epimerase |
| chr2_+_242254679 | 2.28 |
ENST00000428282.1 ENST00000360051.3 |
SEPT2 |
septin 2 |
| chr7_-_30066233 | 2.23 |
ENST00000222803.5 |
FKBP14 |
FK506 binding protein 14, 22 kDa |
| chr16_-_66959429 | 2.21 |
ENST00000420652.1 ENST00000299759.6 |
RRAD |
Ras-related associated with diabetes |
| chr19_+_16187816 | 2.02 |
ENST00000588410.1 |
TPM4 |
tropomyosin 4 |
| chr10_-_128077024 | 2.00 |
ENST00000368679.4 ENST00000368676.4 ENST00000448723.1 |
ADAM12 |
ADAM metallopeptidase domain 12 |
| chr15_-_60690163 | 1.99 |
ENST00000558998.1 ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2 |
annexin A2 |
| chr2_-_20251744 | 1.96 |
ENST00000175091.4 |
LAPTM4A |
lysosomal protein transmembrane 4 alpha |
| chr9_+_35732312 | 1.93 |
ENST00000353704.2 |
CREB3 |
cAMP responsive element binding protein 3 |
| chr11_-_66056596 | 1.92 |
ENST00000471387.2 ENST00000359461.6 ENST00000376901.4 |
YIF1A |
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr17_-_40575535 | 1.91 |
ENST00000357037.5 |
PTRF |
polymerase I and transcript release factor |
| chr5_-_93447333 | 1.90 |
ENST00000395965.3 ENST00000505869.1 ENST00000509163.1 |
FAM172A |
family with sequence similarity 172, member A |
| chr20_-_17662705 | 1.90 |
ENST00000455029.2 |
RRBP1 |
ribosome binding protein 1 |
| chr9_+_114393634 | 1.89 |
ENST00000556107.1 ENST00000374294.3 |
DNAJC25 DNAJC25-GNG10 |
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr4_-_119757239 | 1.85 |
ENST00000280551.6 |
SEC24D |
SEC24 family member D |
| chr4_-_119757322 | 1.85 |
ENST00000379735.5 |
SEC24D |
SEC24 family member D |
| chr7_-_6523755 | 1.83 |
ENST00000436575.1 ENST00000258739.4 |
DAGLB KDELR2 |
diacylglycerol lipase, beta KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr2_+_242254507 | 1.82 |
ENST00000391973.2 |
SEPT2 |
septin 2 |
| chr6_-_83902933 | 1.79 |
ENST00000512866.1 ENST00000510258.1 ENST00000503094.1 ENST00000283977.4 ENST00000513973.1 ENST00000508748.1 |
PGM3 |
phosphoglucomutase 3 |
| chr11_+_207477 | 1.74 |
ENST00000526104.1 |
RIC8A |
RIC8 guanine nucleotide exchange factor A |
| chr3_+_127771212 | 1.74 |
ENST00000243253.3 ENST00000481210.1 |
SEC61A1 |
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr14_-_24664776 | 1.65 |
ENST00000530468.1 ENST00000528010.1 ENST00000396854.4 ENST00000524835.1 ENST00000261789.4 ENST00000525592.1 |
TM9SF1 |
transmembrane 9 superfamily member 1 |
| chr12_-_133405288 | 1.65 |
ENST00000204726.3 |
GOLGA3 |
golgin A3 |
| chr2_-_242254595 | 1.63 |
ENST00000441124.1 ENST00000391976.2 |
HDLBP |
high density lipoprotein binding protein |
| chr11_-_66056478 | 1.57 |
ENST00000431556.2 ENST00000528575.1 |
YIF1A |
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr1_+_145575980 | 1.56 |
ENST00000393045.2 |
PIAS3 |
protein inhibitor of activated STAT, 3 |
| chr1_+_145576007 | 1.55 |
ENST00000369298.1 |
PIAS3 |
protein inhibitor of activated STAT, 3 |
| chr1_+_21835858 | 1.54 |
ENST00000539907.1 ENST00000540617.1 ENST00000374840.3 |
ALPL |
alkaline phosphatase, liver/bone/kidney |
| chr14_-_24664540 | 1.52 |
ENST00000530563.1 ENST00000528895.1 ENST00000528669.1 ENST00000532632.1 |
TM9SF1 |
transmembrane 9 superfamily member 1 |
| chr20_+_44044717 | 1.51 |
ENST00000279036.6 ENST00000279035.9 ENST00000372689.5 ENST00000545755.1 ENST00000341555.5 ENST00000535404.1 ENST00000543458.2 ENST00000432270.1 |
PIGT |
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr2_-_33824382 | 1.49 |
ENST00000238823.8 |
FAM98A |
family with sequence similarity 98, member A |
| chr2_+_85132749 | 1.44 |
ENST00000233143.4 |
TMSB10 |
thymosin beta 10 |
| chr2_-_33824336 | 1.41 |
ENST00000431950.1 ENST00000403368.1 ENST00000441530.2 |
FAM98A |
family with sequence similarity 98, member A |
| chr3_+_33155525 | 1.39 |
ENST00000449224.1 |
CRTAP |
cartilage associated protein |
| chr3_+_50284321 | 1.34 |
ENST00000451956.1 |
GNAI2 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 |
| chr13_+_35516390 | 1.31 |
ENST00000540320.1 ENST00000400445.3 ENST00000310336.4 |
NBEA |
neurobeachin |
| chr11_+_17298297 | 1.27 |
ENST00000529010.1 |
NUCB2 |
nucleobindin 2 |
| chr17_+_8339189 | 1.24 |
ENST00000585098.1 ENST00000380025.4 ENST00000402554.3 ENST00000584866.1 ENST00000582490.1 |
NDEL1 |
nudE neurodevelopment protein 1-like 1 |
| chr1_-_144932464 | 1.23 |
ENST00000479408.2 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr15_+_79603404 | 1.23 |
ENST00000299705.5 |
TMED3 |
transmembrane emp24 protein transport domain containing 3 |
| chr19_+_36606654 | 1.20 |
ENST00000588385.1 ENST00000585746.1 |
TBCB |
tubulin folding cofactor B |
| chr20_+_25228669 | 1.18 |
ENST00000216962.4 |
PYGB |
phosphorylase, glycogen; brain |
| chr6_+_33168637 | 1.18 |
ENST00000374677.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr6_+_33168597 | 1.14 |
ENST00000374675.3 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
| chr18_-_34408902 | 1.13 |
ENST00000593035.1 ENST00000383056.3 ENST00000588909.1 ENST00000590337.1 |
TPGS2 |
tubulin polyglutamylase complex subunit 2 |
| chr14_-_92506371 | 1.13 |
ENST00000267622.4 |
TRIP11 |
thyroid hormone receptor interactor 11 |
| chr1_+_87170577 | 1.09 |
ENST00000482504.1 |
SH3GLB1 |
SH3-domain GRB2-like endophilin B1 |
| chr6_+_83903061 | 1.07 |
ENST00000369724.4 ENST00000539997.1 |
RWDD2A |
RWD domain containing 2A |
| chr11_+_17298255 | 1.06 |
ENST00000531172.1 ENST00000533738.2 ENST00000323688.6 |
NUCB2 |
nucleobindin 2 |
| chr17_+_8339164 | 1.05 |
ENST00000582665.1 ENST00000334527.7 ENST00000299734.7 |
NDEL1 |
nudE neurodevelopment protein 1-like 1 |
| chr18_-_34409116 | 1.00 |
ENST00000334295.4 |
TPGS2 |
tubulin polyglutamylase complex subunit 2 |
| chr11_-_207221 | 0.99 |
ENST00000486280.1 ENST00000332865.6 ENST00000529614.2 ENST00000325147.9 ENST00000410108.1 ENST00000382762.3 |
BET1L |
Bet1 golgi vesicular membrane trafficking protein-like |
| chr17_+_64298944 | 0.96 |
ENST00000413366.3 |
PRKCA |
protein kinase C, alpha |
| chr6_+_132129151 | 0.96 |
ENST00000360971.2 |
ENPP1 |
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr1_-_145039949 | 0.95 |
ENST00000313382.9 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr14_+_102430855 | 0.95 |
ENST00000360184.4 |
DYNC1H1 |
dynein, cytoplasmic 1, heavy chain 1 |
| chr7_-_6523688 | 0.93 |
ENST00000490996.1 |
KDELR2 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr1_-_87379785 | 0.91 |
ENST00000401030.3 ENST00000370554.1 |
SEP15 |
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr16_+_56965960 | 0.87 |
ENST00000439977.2 ENST00000344114.4 ENST00000300302.5 ENST00000379792.2 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr17_+_46048376 | 0.87 |
ENST00000338399.4 |
CDK5RAP3 |
CDK5 regulatory subunit associated protein 3 |
| chr2_-_99952769 | 0.86 |
ENST00000409434.1 ENST00000434323.1 ENST00000264255.3 |
TXNDC9 |
thioredoxin domain containing 9 |
| chr5_-_115177496 | 0.80 |
ENST00000274459.4 ENST00000509910.1 |
ATG12 |
autophagy related 12 |
| chr3_-_156272924 | 0.79 |
ENST00000467789.1 ENST00000265044.2 |
SSR3 |
signal sequence receptor, gamma (translocon-associated protein gamma) |
| chr14_+_50087468 | 0.78 |
ENST00000305386.2 |
MGAT2 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr11_-_68039364 | 0.77 |
ENST00000533310.1 ENST00000304271.6 ENST00000527280.1 |
C11orf24 |
chromosome 11 open reading frame 24 |
| chr22_-_43253189 | 0.74 |
ENST00000437119.2 ENST00000429508.2 ENST00000454099.1 ENST00000263245.5 |
ARFGAP3 |
ADP-ribosylation factor GTPase activating protein 3 |
| chrX_-_41782683 | 0.74 |
ENST00000378163.1 ENST00000378154.1 |
CASK |
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr6_-_33168391 | 0.72 |
ENST00000374685.4 ENST00000413614.2 ENST00000374680.3 |
RXRB |
retinoid X receptor, beta |
| chr19_+_1026566 | 0.69 |
ENST00000348419.3 ENST00000565096.2 ENST00000562958.2 ENST00000562075.2 ENST00000607102.1 |
CNN2 |
calponin 2 |
| chr1_+_26758790 | 0.68 |
ENST00000427245.2 ENST00000525682.2 ENST00000236342.7 ENST00000526219.1 ENST00000374185.3 ENST00000360009.2 |
DHDDS |
dehydrodolichyl diphosphate synthase |
| chrX_-_153059958 | 0.67 |
ENST00000370092.3 ENST00000217901.5 |
IDH3G |
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr4_+_56262115 | 0.66 |
ENST00000506198.1 ENST00000381334.5 ENST00000542052.1 |
TMEM165 |
transmembrane protein 165 |
| chr5_+_112196919 | 0.63 |
ENST00000505459.1 ENST00000282999.3 ENST00000515463.1 |
SRP19 |
signal recognition particle 19kDa |
| chr3_-_139108475 | 0.60 |
ENST00000515006.1 ENST00000513274.1 ENST00000514508.1 ENST00000507777.1 ENST00000512153.1 ENST00000333188.5 |
COPB2 |
coatomer protein complex, subunit beta 2 (beta prime) |
| chr10_-_30348439 | 0.58 |
ENST00000375377.1 |
KIAA1462 |
KIAA1462 |
| chr15_-_75165651 | 0.57 |
ENST00000562363.1 ENST00000564529.1 ENST00000268099.9 |
SCAMP2 |
secretory carrier membrane protein 2 |
| chr5_-_108745689 | 0.56 |
ENST00000361189.2 |
PJA2 |
praja ring finger 2, E3 ubiquitin protein ligase |
| chr1_+_6845384 | 0.55 |
ENST00000303635.7 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr19_+_33182823 | 0.54 |
ENST00000397061.3 |
NUDT19 |
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr6_-_108279369 | 0.52 |
ENST00000369002.4 |
SEC63 |
SEC63 homolog (S. cerevisiae) |
| chrX_-_153059811 | 0.46 |
ENST00000427365.2 ENST00000444450.1 ENST00000370093.1 |
IDH3G |
isocitrate dehydrogenase 3 (NAD+) gamma |
| chrX_+_153060090 | 0.44 |
ENST00000370086.3 ENST00000370085.3 |
SSR4 |
signal sequence receptor, delta |
| chr10_+_104678102 | 0.43 |
ENST00000433628.2 |
CNNM2 |
cyclin M2 |
| chr4_+_26322185 | 0.43 |
ENST00000361572.6 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
| chr11_-_62599505 | 0.43 |
ENST00000377897.4 ENST00000394690.1 ENST00000541317.1 ENST00000294179.3 |
STX5 |
syntaxin 5 |
| chr1_-_87380002 | 0.41 |
ENST00000331835.5 |
SEP15 |
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr11_+_118443098 | 0.41 |
ENST00000392859.3 ENST00000359415.4 ENST00000534182.2 ENST00000264028.4 |
ARCN1 |
archain 1 |
| chr3_-_10362725 | 0.40 |
ENST00000397109.3 ENST00000428626.1 ENST00000445064.1 ENST00000431352.1 ENST00000397117.1 ENST00000337354.4 ENST00000383801.2 ENST00000432213.1 ENST00000350697.3 |
SEC13 |
SEC13 homolog (S. cerevisiae) |
| chr21_-_18985230 | 0.39 |
ENST00000457956.1 ENST00000348354.6 |
BTG3 |
BTG family, member 3 |
| chr4_+_190992087 | 0.35 |
ENST00000553598.1 |
DUX4L7 |
double homeobox 4 like 7 |
| chr10_+_112257596 | 0.35 |
ENST00000369583.3 |
DUSP5 |
dual specificity phosphatase 5 |
| chr8_-_124054362 | 0.34 |
ENST00000405944.3 |
DERL1 |
derlin 1 |
| chr1_+_207669573 | 0.31 |
ENST00000400960.2 ENST00000534202.1 |
CR1 |
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr3_-_105588231 | 0.31 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr2_+_27255806 | 0.31 |
ENST00000238788.9 ENST00000404032.3 |
TMEM214 |
transmembrane protein 214 |
| chr18_-_34408693 | 0.30 |
ENST00000587382.1 ENST00000589049.1 ENST00000587129.1 |
TPGS2 |
tubulin polyglutamylase complex subunit 2 |
| chr8_+_105235572 | 0.29 |
ENST00000523362.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr8_+_67624653 | 0.28 |
ENST00000521198.2 |
SGK3 |
serum/glucocorticoid regulated kinase family, member 3 |
| chr12_-_109125285 | 0.27 |
ENST00000552871.1 ENST00000261401.3 |
CORO1C |
coronin, actin binding protein, 1C |
| chr19_+_17186577 | 0.26 |
ENST00000595618.1 ENST00000594824.1 |
MYO9B |
myosin IXB |
| chr19_-_4670345 | 0.26 |
ENST00000599630.1 ENST00000262947.3 |
C19orf10 |
chromosome 19 open reading frame 10 |
| chr19_-_54619006 | 0.26 |
ENST00000391759.1 |
TFPT |
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr5_+_126853301 | 0.25 |
ENST00000296666.8 ENST00000442138.2 ENST00000512635.2 |
PRRC1 |
proline-rich coiled-coil 1 |
| chr19_+_50353944 | 0.24 |
ENST00000594151.1 ENST00000600603.1 ENST00000601638.1 ENST00000221557.9 |
PTOV1 |
prostate tumor overexpressed 1 |
| chr11_+_64851666 | 0.23 |
ENST00000525509.1 ENST00000294258.3 ENST00000526334.1 |
ZFPL1 |
zinc finger protein-like 1 |
| chr10_+_104678032 | 0.22 |
ENST00000369878.4 ENST00000369875.3 |
CNNM2 |
cyclin M2 |
| chr4_+_26322409 | 0.22 |
ENST00000514807.1 ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
| chr1_-_144932316 | 0.22 |
ENST00000313431.9 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chrX_-_41782249 | 0.21 |
ENST00000442742.2 ENST00000421587.2 ENST00000378166.4 ENST00000318588.9 ENST00000361962.4 |
CASK |
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chrX_+_153059608 | 0.21 |
ENST00000370087.1 |
SSR4 |
signal sequence receptor, delta |
| chr12_+_99038919 | 0.20 |
ENST00000551964.1 |
APAF1 |
apoptotic peptidase activating factor 1 |
| chr3_-_105587879 | 0.18 |
ENST00000264122.4 ENST00000403724.1 ENST00000405772.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr20_+_57267669 | 0.16 |
ENST00000356091.6 |
NPEPL1 |
aminopeptidase-like 1 |
| chr20_-_8000426 | 0.15 |
ENST00000527925.1 ENST00000246024.2 |
TMX4 |
thioredoxin-related transmembrane protein 4 |
| chr19_+_50354393 | 0.15 |
ENST00000391842.1 |
PTOV1 |
prostate tumor overexpressed 1 |
| chr11_-_59383617 | 0.15 |
ENST00000263847.1 |
OSBP |
oxysterol binding protein |
| chr1_+_207669613 | 0.14 |
ENST00000367049.4 ENST00000529814.1 |
CR1 |
complement component (3b/4b) receptor 1 (Knops blood group) |
| chr19_-_19030157 | 0.13 |
ENST00000349893.4 ENST00000351079.4 ENST00000600932.1 ENST00000262812.4 |
COPE |
coatomer protein complex, subunit epsilon |
| chr19_-_54618650 | 0.12 |
ENST00000391757.1 |
TFPT |
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr8_-_124054484 | 0.11 |
ENST00000419562.2 |
DERL1 |
derlin 1 |
| chr4_+_76649797 | 0.09 |
ENST00000538159.1 ENST00000514213.2 |
USO1 |
USO1 vesicle transport factor |
| chr19_+_47104493 | 0.06 |
ENST00000291295.9 ENST00000597743.1 |
CALM3 |
calmodulin 3 (phosphorylase kinase, delta) |
| chr9_-_35732362 | 0.05 |
ENST00000314888.9 ENST00000540444.1 |
TLN1 |
talin 1 |
| chr3_+_132378741 | 0.04 |
ENST00000493720.2 |
UBA5 |
ubiquitin-like modifier activating enzyme 5 |
| chr13_+_26042960 | 0.04 |
ENST00000255283.8 |
ATP8A2 |
ATPase, aminophospholipid transporter, class I, type 8A, member 2 |
| chr21_-_18985158 | 0.04 |
ENST00000339775.6 |
BTG3 |
BTG family, member 3 |
| chr22_-_20850128 | 0.04 |
ENST00000328879.4 |
KLHL22 |
kelch-like family member 22 |
| chr3_-_128369643 | 0.04 |
ENST00000296255.3 |
RPN1 |
ribophorin I |
| chr11_-_64646086 | 0.03 |
ENST00000320631.3 |
EHD1 |
EH-domain containing 1 |
| chr1_-_45988542 | 0.03 |
ENST00000424390.1 |
PRDX1 |
peroxiredoxin 1 |
| chr11_-_62414070 | 0.00 |
ENST00000540933.1 ENST00000346178.4 ENST00000356638.3 ENST00000534779.1 ENST00000525994.1 |
GANAB |
glucosidase, alpha; neutral AB |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 21.0 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 1.2 | 5.8 | GO:0044354 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.8 | 2.3 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.6 | 7.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.5 | 1.6 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.5 | 1.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.4 | 2.9 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.4 | 1.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) outer acrosomal membrane(GO:0002081) |
| 0.3 | 4.1 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.3 | 1.5 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.2 | 2.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 2.7 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.2 | 4.6 | GO:0042599 | lamellar body(GO:0042599) |
| 0.2 | 2.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 3.5 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 0.9 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.8 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 4.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 2.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 3.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 3.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 5.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.3 | GO:0036502 | Derlin-1-VIMP complex(GO:0036502) |
| 0.1 | 0.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 7.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 6.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 9.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 3.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.9 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 2.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 2.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 9.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 4.7 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.7 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.0 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 2.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 21.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 5.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 3.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 7.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 3.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 4.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 4.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 9.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 2.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 8.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.9 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 5.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 21.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.3 | 7.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.3 | 6.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 7.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 10.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 3.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 2.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 9.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 1.9 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 2.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.8 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.9 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 3.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 2.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.2 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.9 | 21.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 1.1 | 8.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 1.0 | 5.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.8 | 2.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.6 | 2.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.5 | 1.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.4 | 6.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.4 | 9.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 1.0 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.3 | 1.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.3 | 1.8 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 1.2 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 1.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 1.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 9.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 5.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 0.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 0.9 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.2 | 0.5 | GO:0001861 | complement component C4b receptor activity(GO:0001861) complement component C3b receptor activity(GO:0004877) |
| 0.1 | 3.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.8 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.1 | 2.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.0 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.6 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 2.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.9 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 3.3 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 0.7 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 1.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.9 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.7 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.5 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 2.9 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 1.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 2.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 2.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 6.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 8.4 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0032804 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 1.9 | 7.4 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 1.4 | 21.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 1.1 | 3.3 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 1.1 | 3.3 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.9 | 9.8 | GO:0031584 | activation of phospholipase D activity(GO:0031584) apical protein localization(GO:0045176) |
| 0.8 | 8.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.7 | 2.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.6 | 7.7 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.5 | 6.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.5 | 1.5 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.5 | 5.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.5 | 1.4 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.4 | 1.9 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.4 | 1.8 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.3 | 1.7 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.3 | 0.7 | GO:0072507 | magnesium ion homeostasis(GO:0010960) divalent inorganic cation homeostasis(GO:0072507) |
| 0.3 | 1.0 | GO:2000705 | histone H3-T6 phosphorylation(GO:0035408) dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.3 | 3.1 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.3 | 1.1 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.3 | 3.9 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.3 | 2.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.3 | 1.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 0.7 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.2 | 0.9 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.2 | 0.6 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.2 | 2.7 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.2 | 3.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.2 | 2.5 | GO:0048843 | positive regulation of axon extension involved in axon guidance(GO:0048842) negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.2 | 0.9 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 | 1.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 1.7 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.2 | 0.5 | GO:0030451 | regulation of complement activation, alternative pathway(GO:0030451) negative regulation of complement activation, alternative pathway(GO:0045957) |
| 0.1 | 1.0 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 1.1 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 2.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.4 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 | 2.4 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.7 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.1 | 1.1 | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.1 | 1.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.6 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.1 | 0.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 1.3 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.9 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.9 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.3 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.5 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 2.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 2.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.8 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.8 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 3.3 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 2.0 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.3 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 4.1 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 2.0 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 10.7 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.0 | GO:0003011 | involuntary skeletal muscle contraction(GO:0003011) |
| 0.0 | 1.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.7 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.6 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.6 | GO:0007155 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.0 | 0.6 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 2.9 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 0.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 3.1 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.6 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |