Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
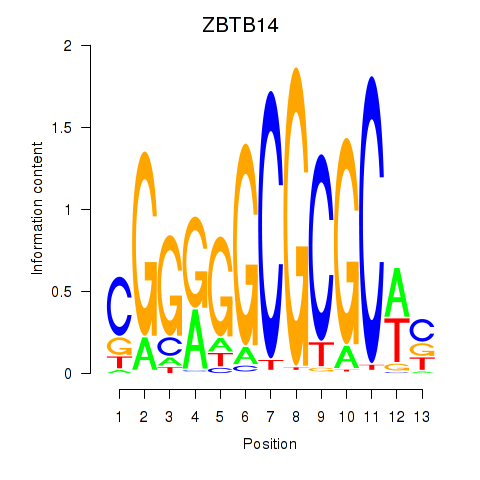
Results for ZBTB14
Z-value: 0.74
Transcription factors associated with ZBTB14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB14
|
ENSG00000198081.6 | ZBTB14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB14 | hg19_v2_chr18_-_5296001_5296045 | 0.55 | 2.6e-02 | Click! |
Activity profile of ZBTB14 motif
Sorted Z-values of ZBTB14 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB14
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_31744031 | 0.83 |
ENST00000389082.5 |
DENND5B |
DENN/MADD domain containing 5B |
| chr10_+_11060004 | 0.82 |
ENST00000542579.1 ENST00000399850.3 ENST00000417956.2 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr3_+_142838091 | 0.78 |
ENST00000309575.3 |
CHST2 |
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
| chr7_-_55640176 | 0.76 |
ENST00000285279.5 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr6_-_90121789 | 0.73 |
ENST00000359203.3 |
RRAGD |
Ras-related GTP binding D |
| chr1_+_111682827 | 0.70 |
ENST00000357172.4 |
CEPT1 |
choline/ethanolamine phosphotransferase 1 |
| chr9_-_92112953 | 0.69 |
ENST00000339861.4 ENST00000422704.2 ENST00000455551.2 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
| chr1_+_236305826 | 0.68 |
ENST00000366592.3 ENST00000366591.4 |
GPR137B |
G protein-coupled receptor 137B |
| chr5_-_88179302 | 0.68 |
ENST00000504921.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr1_+_174128639 | 0.66 |
ENST00000251507.4 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr10_+_11059826 | 0.65 |
ENST00000450189.1 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr15_-_38856836 | 0.64 |
ENST00000450598.2 ENST00000559830.1 ENST00000558164.1 ENST00000310803.5 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
| chr7_+_98246588 | 0.61 |
ENST00000265634.3 |
NPTX2 |
neuronal pentraxin II |
| chr9_+_71320596 | 0.59 |
ENST00000265382.3 |
PIP5K1B |
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr11_+_17741111 | 0.58 |
ENST00000250003.3 |
MYOD1 |
myogenic differentiation 1 |
| chr6_+_135502408 | 0.57 |
ENST00000341911.5 ENST00000442647.2 ENST00000316528.8 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr20_+_9049682 | 0.57 |
ENST00000334005.3 ENST00000378473.3 |
PLCB4 |
phospholipase C, beta 4 |
| chr9_+_137218362 | 0.57 |
ENST00000481739.1 |
RXRA |
retinoid X receptor, alpha |
| chr6_+_135502466 | 0.54 |
ENST00000367814.4 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr1_+_150122034 | 0.52 |
ENST00000025469.6 ENST00000369124.4 |
PLEKHO1 |
pleckstrin homology domain containing, family O member 1 |
| chr5_-_88179017 | 0.50 |
ENST00000514028.1 ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr22_+_24129138 | 0.49 |
ENST00000417137.1 ENST00000344921.6 ENST00000263121.7 ENST00000407422.3 ENST00000407082.3 |
SMARCB1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 |
| chr14_+_71108460 | 0.48 |
ENST00000256367.2 |
TTC9 |
tetratricopeptide repeat domain 9 |
| chr5_+_61602055 | 0.47 |
ENST00000381103.2 |
KIF2A |
kinesin heavy chain member 2A |
| chr13_-_36705425 | 0.47 |
ENST00000255448.4 ENST00000360631.3 ENST00000379892.4 |
DCLK1 |
doublecortin-like kinase 1 |
| chrX_-_107019181 | 0.47 |
ENST00000315660.4 ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr5_-_88178964 | 0.45 |
ENST00000513252.1 ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr2_+_207308539 | 0.45 |
ENST00000374416.1 ENST00000374415.3 |
ADAM23 |
ADAM metallopeptidase domain 23 |
| chr16_+_30194916 | 0.44 |
ENST00000570045.1 ENST00000565497.1 ENST00000570244.1 |
CORO1A |
coronin, actin binding protein, 1A |
| chr3_-_107809816 | 0.44 |
ENST00000361309.5 ENST00000355354.7 |
CD47 |
CD47 molecule |
| chr19_-_46974664 | 0.42 |
ENST00000438932.2 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr19_-_46974741 | 0.42 |
ENST00000313683.10 ENST00000602246.1 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr17_+_5185552 | 0.40 |
ENST00000262477.6 ENST00000408982.2 ENST00000575991.1 ENST00000537505.1 ENST00000546142.2 |
RABEP1 |
rabaptin, RAB GTPase binding effector protein 1 |
| chr20_+_57466629 | 0.39 |
ENST00000371081.1 ENST00000338783.6 |
GNAS |
GNAS complex locus |
| chr9_+_71320557 | 0.39 |
ENST00000541509.1 |
PIP5K1B |
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr9_-_115095883 | 0.39 |
ENST00000450374.1 ENST00000374255.2 ENST00000334318.6 ENST00000374257.1 |
PTBP3 |
polypyrimidine tract binding protein 3 |
| chr20_+_57430162 | 0.36 |
ENST00000450130.1 ENST00000349036.3 ENST00000423897.1 |
GNAS |
GNAS complex locus |
| chr11_-_33891362 | 0.36 |
ENST00000395833.3 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr3_-_141868293 | 0.36 |
ENST00000317104.7 ENST00000494358.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr14_+_60715928 | 0.36 |
ENST00000395076.4 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr17_-_40428359 | 0.35 |
ENST00000293328.3 |
STAT5B |
signal transducer and activator of transcription 5B |
| chr1_+_210502238 | 0.34 |
ENST00000545154.1 ENST00000537898.1 ENST00000391905.3 ENST00000545781.1 ENST00000261458.3 ENST00000308852.6 |
HHAT |
hedgehog acyltransferase |
| chr14_+_60716159 | 0.34 |
ENST00000325658.3 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr2_-_86564776 | 0.34 |
ENST00000165698.5 ENST00000541910.1 ENST00000535845.1 |
REEP1 |
receptor accessory protein 1 |
| chr20_-_62710832 | 0.33 |
ENST00000395042.1 |
RGS19 |
regulator of G-protein signaling 19 |
| chr1_-_40367668 | 0.33 |
ENST00000397332.2 ENST00000429311.1 |
MYCL |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr2_+_234263120 | 0.32 |
ENST00000264057.2 ENST00000427930.1 |
DGKD |
diacylglycerol kinase, delta 130kDa |
| chr17_+_43299241 | 0.31 |
ENST00000328118.3 |
FMNL1 |
formin-like 1 |
| chr12_-_31479045 | 0.31 |
ENST00000539409.1 ENST00000395766.1 |
FAM60A |
family with sequence similarity 60, member A |
| chr20_-_56284816 | 0.31 |
ENST00000395819.3 ENST00000341744.3 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr2_-_175547571 | 0.31 |
ENST00000409415.3 ENST00000359761.3 ENST00000272746.5 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr18_-_5296001 | 0.30 |
ENST00000357006.4 |
ZBTB14 |
zinc finger and BTB domain containing 14 |
| chr2_+_71693812 | 0.29 |
ENST00000409651.1 ENST00000394120.2 ENST00000409744.1 ENST00000409366.1 ENST00000410020.3 ENST00000410041.1 |
DYSF |
dysferlin |
| chr8_-_90996459 | 0.29 |
ENST00000517337.1 ENST00000409330.1 |
NBN |
nibrin |
| chr3_+_133293278 | 0.29 |
ENST00000508481.1 ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3 |
CDV3 homolog (mouse) |
| chr10_+_88854926 | 0.29 |
ENST00000298784.1 ENST00000298786.4 |
FAM35A |
family with sequence similarity 35, member A |
| chr11_-_46142948 | 0.29 |
ENST00000257821.4 |
PHF21A |
PHD finger protein 21A |
| chrX_+_11776410 | 0.29 |
ENST00000361672.2 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
| chr2_-_225907150 | 0.29 |
ENST00000258390.7 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr18_-_11148587 | 0.29 |
ENST00000302079.6 ENST00000580640.1 ENST00000503781.3 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
| chr21_-_46237883 | 0.28 |
ENST00000397893.3 |
SUMO3 |
small ubiquitin-like modifier 3 |
| chr9_+_117373486 | 0.28 |
ENST00000288502.4 ENST00000374049.4 |
C9orf91 |
chromosome 9 open reading frame 91 |
| chr17_+_75137460 | 0.28 |
ENST00000587820.1 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
| chr6_-_43337180 | 0.28 |
ENST00000318149.3 ENST00000361428.2 |
ZNF318 |
zinc finger protein 318 |
| chr15_+_75074410 | 0.28 |
ENST00000439220.2 |
CSK |
c-src tyrosine kinase |
| chrX_-_3631635 | 0.28 |
ENST00000262848.5 |
PRKX |
protein kinase, X-linked |
| chr3_-_197476560 | 0.27 |
ENST00000273582.5 |
KIAA0226 |
KIAA0226 |
| chr2_-_242088850 | 0.27 |
ENST00000358649.4 ENST00000452907.1 ENST00000403638.3 ENST00000539818.1 ENST00000415234.1 ENST00000234040.4 |
PASK |
PAS domain containing serine/threonine kinase |
| chr14_+_103243813 | 0.27 |
ENST00000560371.1 ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3 |
TNF receptor-associated factor 3 |
| chr16_+_6533380 | 0.27 |
ENST00000552089.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr8_-_90996837 | 0.27 |
ENST00000519426.1 ENST00000265433.3 |
NBN |
nibrin |
| chr5_+_176560742 | 0.27 |
ENST00000439151.2 |
NSD1 |
nuclear receptor binding SET domain protein 1 |
| chr21_+_44394742 | 0.27 |
ENST00000432907.2 |
PKNOX1 |
PBX/knotted 1 homeobox 1 |
| chr2_+_207308220 | 0.27 |
ENST00000264377.3 |
ADAM23 |
ADAM metallopeptidase domain 23 |
| chr5_-_94620239 | 0.27 |
ENST00000515393.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr17_+_43299156 | 0.26 |
ENST00000331495.3 |
FMNL1 |
formin-like 1 |
| chr7_+_150756657 | 0.26 |
ENST00000413384.2 |
SLC4A2 |
solute carrier family 4 (anion exchanger), member 2 |
| chr15_+_91643442 | 0.26 |
ENST00000394232.1 |
SV2B |
synaptic vesicle glycoprotein 2B |
| chr1_-_151431647 | 0.26 |
ENST00000368863.2 ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ |
pogo transposable element with ZNF domain |
| chr3_+_133292759 | 0.26 |
ENST00000431519.2 |
CDV3 |
CDV3 homolog (mouse) |
| chr8_+_61591337 | 0.26 |
ENST00000423902.2 |
CHD7 |
chromodomain helicase DNA binding protein 7 |
| chr1_-_29508499 | 0.24 |
ENST00000373795.4 |
SRSF4 |
serine/arginine-rich splicing factor 4 |
| chr8_+_142402089 | 0.24 |
ENST00000521578.1 ENST00000520105.1 ENST00000523147.1 |
PTP4A3 |
protein tyrosine phosphatase type IVA, member 3 |
| chr11_+_65029421 | 0.24 |
ENST00000541089.1 |
POLA2 |
polymerase (DNA directed), alpha 2, accessory subunit |
| chr14_-_102553371 | 0.24 |
ENST00000553585.1 ENST00000216281.8 |
HSP90AA1 |
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr4_+_75858318 | 0.24 |
ENST00000307428.7 |
PARM1 |
prostate androgen-regulated mucin-like protein 1 |
| chr21_+_38071430 | 0.24 |
ENST00000290399.6 |
SIM2 |
single-minded family bHLH transcription factor 2 |
| chr4_+_75858290 | 0.24 |
ENST00000513238.1 |
PARM1 |
prostate androgen-regulated mucin-like protein 1 |
| chr17_+_30264014 | 0.23 |
ENST00000322652.5 ENST00000580398.1 |
SUZ12 |
SUZ12 polycomb repressive complex 2 subunit |
| chr12_+_111843749 | 0.23 |
ENST00000341259.2 |
SH2B3 |
SH2B adaptor protein 3 |
| chr6_-_86352982 | 0.23 |
ENST00000369622.3 |
SYNCRIP |
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chrX_+_11776278 | 0.23 |
ENST00000312196.4 ENST00000337339.2 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
| chr3_-_49449350 | 0.23 |
ENST00000454011.2 ENST00000445425.1 ENST00000422781.1 |
RHOA |
ras homolog family member A |
| chr5_+_102595119 | 0.23 |
ENST00000510890.1 |
C5orf30 |
chromosome 5 open reading frame 30 |
| chr5_+_176561129 | 0.23 |
ENST00000511258.1 ENST00000347982.4 |
NSD1 |
nuclear receptor binding SET domain protein 1 |
| chr17_+_35766965 | 0.23 |
ENST00000394395.2 ENST00000589153.1 ENST00000586023.1 |
TADA2A |
transcriptional adaptor 2A |
| chr2_+_79740118 | 0.23 |
ENST00000496558.1 ENST00000451966.1 |
CTNNA2 |
catenin (cadherin-associated protein), alpha 2 |
| chr6_+_135502501 | 0.22 |
ENST00000527615.1 ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr4_+_41362796 | 0.22 |
ENST00000508501.1 ENST00000512946.1 ENST00000313860.7 ENST00000512632.1 ENST00000512820.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr1_-_29508321 | 0.22 |
ENST00000546138.1 |
SRSF4 |
serine/arginine-rich splicing factor 4 |
| chr3_-_133614597 | 0.22 |
ENST00000285208.4 ENST00000460865.3 |
RAB6B |
RAB6B, member RAS oncogene family |
| chr17_-_31204124 | 0.22 |
ENST00000579584.1 ENST00000318217.5 ENST00000583621.1 |
MYO1D |
myosin ID |
| chr3_+_133292851 | 0.22 |
ENST00000503932.1 |
CDV3 |
CDV3 homolog (mouse) |
| chr13_-_95953589 | 0.22 |
ENST00000538287.1 ENST00000376887.4 ENST00000412704.1 ENST00000536256.1 ENST00000431522.1 |
ABCC4 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr7_-_5569588 | 0.22 |
ENST00000417101.1 |
ACTB |
actin, beta |
| chr6_-_136610911 | 0.22 |
ENST00000530767.1 ENST00000527759.1 ENST00000527536.1 ENST00000529826.1 ENST00000531224.1 ENST00000353331.4 |
BCLAF1 |
BCL2-associated transcription factor 1 |
| chr6_+_108881012 | 0.21 |
ENST00000343882.6 |
FOXO3 |
forkhead box O3 |
| chr6_+_12012536 | 0.21 |
ENST00000379388.2 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
| chr6_+_24495067 | 0.21 |
ENST00000357578.3 ENST00000546278.1 ENST00000491546.1 |
ALDH5A1 |
aldehyde dehydrogenase 5 family, member A1 |
| chr13_-_49107303 | 0.21 |
ENST00000344532.3 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr1_-_32801825 | 0.21 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chr18_+_77155942 | 0.21 |
ENST00000397790.2 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr14_-_105635090 | 0.21 |
ENST00000331782.3 ENST00000347004.2 |
JAG2 |
jagged 2 |
| chr18_+_74534479 | 0.21 |
ENST00000320610.9 |
ZNF236 |
zinc finger protein 236 |
| chr10_+_26986582 | 0.21 |
ENST00000376215.5 ENST00000376203.5 |
PDSS1 |
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr11_+_65837907 | 0.20 |
ENST00000320580.4 |
PACS1 |
phosphofurin acidic cluster sorting protein 1 |
| chr2_+_28113583 | 0.20 |
ENST00000344773.2 ENST00000379624.1 ENST00000342045.2 ENST00000379632.2 ENST00000361704.2 |
BRE |
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr15_-_70388943 | 0.20 |
ENST00000559048.1 ENST00000560939.1 ENST00000440567.3 ENST00000557907.1 ENST00000558379.1 ENST00000451782.2 ENST00000559929.1 |
TLE3 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr17_-_42276574 | 0.20 |
ENST00000589805.1 |
ATXN7L3 |
ataxin 7-like 3 |
| chr7_-_5570229 | 0.20 |
ENST00000331789.5 |
ACTB |
actin, beta |
| chr6_-_31864977 | 0.20 |
ENST00000395728.3 ENST00000375528.4 |
EHMT2 |
euchromatic histone-lysine N-methyltransferase 2 |
| chr21_-_46238034 | 0.20 |
ENST00000332859.6 |
SUMO3 |
small ubiquitin-like modifier 3 |
| chr17_+_80416482 | 0.20 |
ENST00000309794.11 ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF |
nuclear prelamin A recognition factor |
| chr22_-_39268308 | 0.20 |
ENST00000407418.3 |
CBX6 |
chromobox homolog 6 |
| chr20_+_42295745 | 0.20 |
ENST00000396863.4 ENST00000217026.4 |
MYBL2 |
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr19_-_49314269 | 0.20 |
ENST00000545387.2 ENST00000316273.6 ENST00000402551.1 ENST00000598162.1 ENST00000599246.1 |
BCAT2 |
branched chain amino-acid transaminase 2, mitochondrial |
| chr9_-_23821273 | 0.20 |
ENST00000380110.4 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chrX_-_109561294 | 0.19 |
ENST00000372059.2 ENST00000262844.5 |
AMMECR1 |
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr19_-_49314169 | 0.19 |
ENST00000597011.1 ENST00000601681.1 |
BCAT2 |
branched chain amino-acid transaminase 2, mitochondrial |
| chr14_+_64319669 | 0.19 |
ENST00000358025.3 ENST00000357395.3 ENST00000344113.4 ENST00000341472.5 ENST00000356081.3 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
| chr6_+_36410762 | 0.19 |
ENST00000483557.1 ENST00000498267.1 ENST00000544295.1 ENST00000449081.2 ENST00000536244.1 ENST00000460983.1 |
KCTD20 |
potassium channel tetramerization domain containing 20 |
| chr8_+_28351707 | 0.19 |
ENST00000537916.1 ENST00000523546.1 ENST00000240093.3 |
FZD3 |
frizzled family receptor 3 |
| chr3_-_127541679 | 0.19 |
ENST00000265052.5 |
MGLL |
monoglyceride lipase |
| chr9_-_34589700 | 0.19 |
ENST00000351266.4 |
CNTFR |
ciliary neurotrophic factor receptor |
| chr16_-_62070305 | 0.19 |
ENST00000584337.1 |
CDH8 |
cadherin 8, type 2 |
| chr5_+_65440032 | 0.19 |
ENST00000334121.6 |
SREK1 |
splicing regulatory glutamine/lysine-rich protein 1 |
| chr7_+_50344289 | 0.19 |
ENST00000413698.1 ENST00000359197.5 ENST00000331340.3 ENST00000357364.4 ENST00000343574.5 ENST00000349824.4 ENST00000346667.4 ENST00000440768.2 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
| chr9_+_114659046 | 0.18 |
ENST00000374279.3 |
UGCG |
UDP-glucose ceramide glucosyltransferase |
| chr1_-_236445251 | 0.18 |
ENST00000354619.5 ENST00000327333.8 |
ERO1LB |
ERO1-like beta (S. cerevisiae) |
| chr2_+_204193129 | 0.18 |
ENST00000417864.1 |
ABI2 |
abl-interactor 2 |
| chr21_+_44394620 | 0.18 |
ENST00000291547.5 |
PKNOX1 |
PBX/knotted 1 homeobox 1 |
| chr2_-_101034070 | 0.18 |
ENST00000264249.3 |
CHST10 |
carbohydrate sulfotransferase 10 |
| chr17_+_12692774 | 0.18 |
ENST00000379672.5 ENST00000340825.3 |
ARHGAP44 |
Rho GTPase activating protein 44 |
| chrX_-_153363125 | 0.18 |
ENST00000407218.1 ENST00000453960.2 |
MECP2 |
methyl CpG binding protein 2 (Rett syndrome) |
| chr1_+_218519577 | 0.18 |
ENST00000366930.4 ENST00000366929.4 |
TGFB2 |
transforming growth factor, beta 2 |
| chr3_-_133614421 | 0.18 |
ENST00000543906.1 |
RAB6B |
RAB6B, member RAS oncogene family |
| chr17_-_4269920 | 0.18 |
ENST00000572484.1 |
UBE2G1 |
ubiquitin-conjugating enzyme E2G 1 |
| chr12_-_109251345 | 0.18 |
ENST00000360239.3 ENST00000326495.5 ENST00000551165.1 |
SSH1 |
slingshot protein phosphatase 1 |
| chr16_+_29817399 | 0.17 |
ENST00000545521.1 |
MAZ |
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr16_+_19179549 | 0.17 |
ENST00000355377.2 ENST00000568115.1 |
SYT17 |
synaptotagmin XVII |
| chr3_+_147127142 | 0.17 |
ENST00000282928.4 |
ZIC1 |
Zic family member 1 |
| chr22_+_25960786 | 0.17 |
ENST00000324198.6 |
ADRBK2 |
adrenergic, beta, receptor kinase 2 |
| chr11_-_46142615 | 0.17 |
ENST00000529734.1 ENST00000323180.6 |
PHF21A |
PHD finger protein 21A |
| chr20_+_46130671 | 0.17 |
ENST00000371998.3 ENST00000371997.3 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr17_+_21279509 | 0.17 |
ENST00000583088.1 |
KCNJ12 |
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr16_+_56965960 | 0.17 |
ENST00000439977.2 ENST00000344114.4 ENST00000300302.5 ENST00000379792.2 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr9_-_115095851 | 0.17 |
ENST00000343327.2 |
PTBP3 |
polypyrimidine tract binding protein 3 |
| chr7_+_155250824 | 0.17 |
ENST00000297375.4 |
EN2 |
engrailed homeobox 2 |
| chr11_-_78128811 | 0.16 |
ENST00000530915.1 ENST00000361507.4 |
GAB2 |
GRB2-associated binding protein 2 |
| chr14_+_90422239 | 0.16 |
ENST00000393452.3 ENST00000554180.1 ENST00000393454.2 ENST00000553617.1 ENST00000335725.4 ENST00000357382.3 ENST00000556867.1 ENST00000553527.1 |
TDP1 |
tyrosyl-DNA phosphodiesterase 1 |
| chr3_+_14444063 | 0.16 |
ENST00000454876.2 ENST00000360861.3 ENST00000416216.2 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chrX_+_135579670 | 0.16 |
ENST00000218364.4 |
HTATSF1 |
HIV-1 Tat specific factor 1 |
| chr11_+_65029233 | 0.16 |
ENST00000265465.3 |
POLA2 |
polymerase (DNA directed), alpha 2, accessory subunit |
| chr7_+_74072288 | 0.16 |
ENST00000443166.1 |
GTF2I |
general transcription factor IIi |
| chr4_+_41258786 | 0.16 |
ENST00000503431.1 ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr11_+_125495862 | 0.16 |
ENST00000428830.2 ENST00000544373.1 ENST00000527013.1 ENST00000526937.1 ENST00000534685.1 |
CHEK1 |
checkpoint kinase 1 |
| chr17_+_35767301 | 0.16 |
ENST00000225396.6 ENST00000417170.1 ENST00000590005.1 ENST00000590957.1 |
TADA2A |
transcriptional adaptor 2A |
| chr4_-_5894777 | 0.16 |
ENST00000324989.7 |
CRMP1 |
collapsin response mediator protein 1 |
| chr3_-_127542051 | 0.16 |
ENST00000398104.1 |
MGLL |
monoglyceride lipase |
| chr3_-_127542021 | 0.16 |
ENST00000434178.2 |
MGLL |
monoglyceride lipase |
| chr19_+_16435625 | 0.16 |
ENST00000248071.5 ENST00000592003.1 |
KLF2 |
Kruppel-like factor 2 |
| chr5_-_133340326 | 0.16 |
ENST00000425992.1 ENST00000395044.3 ENST00000395047.2 |
VDAC1 |
voltage-dependent anion channel 1 |
| chr7_+_73498118 | 0.16 |
ENST00000336180.2 |
LIMK1 |
LIM domain kinase 1 |
| chr1_+_111682058 | 0.16 |
ENST00000545121.1 |
CEPT1 |
choline/ethanolamine phosphotransferase 1 |
| chr8_+_110552337 | 0.16 |
ENST00000337573.5 |
EBAG9 |
estrogen receptor binding site associated, antigen, 9 |
| chr20_+_46130619 | 0.16 |
ENST00000372004.3 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr8_+_22298578 | 0.16 |
ENST00000240139.5 ENST00000289963.8 ENST00000397775.3 |
PPP3CC |
protein phosphatase 3, catalytic subunit, gamma isozyme |
| chr13_+_114238997 | 0.16 |
ENST00000538138.1 ENST00000375370.5 |
TFDP1 |
transcription factor Dp-1 |
| chr12_-_49351148 | 0.16 |
ENST00000398092.4 ENST00000539611.1 |
RP11-302B13.5 ARF3 |
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr22_+_39853258 | 0.15 |
ENST00000341184.6 |
MGAT3 |
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr14_-_90085458 | 0.15 |
ENST00000345097.4 ENST00000555855.1 ENST00000555353.1 |
FOXN3 |
forkhead box N3 |
| chr8_+_96145974 | 0.15 |
ENST00000315367.3 |
PLEKHF2 |
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr2_+_30454390 | 0.15 |
ENST00000395323.3 ENST00000406087.1 ENST00000404397.1 |
LBH |
limb bud and heart development |
| chrX_+_38420623 | 0.15 |
ENST00000378482.2 |
TSPAN7 |
tetraspanin 7 |
| chr20_+_46130601 | 0.15 |
ENST00000341724.6 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr1_+_165797024 | 0.15 |
ENST00000372212.4 |
UCK2 |
uridine-cytidine kinase 2 |
| chr7_-_45960850 | 0.15 |
ENST00000381083.4 ENST00000381086.5 ENST00000275521.6 |
IGFBP3 |
insulin-like growth factor binding protein 3 |
| chr8_-_74659068 | 0.15 |
ENST00000523558.1 ENST00000521210.1 ENST00000355780.5 ENST00000524104.1 ENST00000521736.1 ENST00000521447.1 ENST00000517542.1 ENST00000521451.1 ENST00000521419.1 ENST00000518502.1 ENST00000524300.1 |
STAU2 |
staufen double-stranded RNA binding protein 2 |
| chr7_+_87563557 | 0.15 |
ENST00000439864.1 ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22 |
ADAM metallopeptidase domain 22 |
| chr15_-_70388599 | 0.15 |
ENST00000560996.1 ENST00000558201.1 |
TLE3 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr1_-_54303949 | 0.15 |
ENST00000234725.8 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr21_+_35445827 | 0.15 |
ENST00000608209.1 ENST00000381151.3 |
SLC5A3 SLC5A3 |
sodium/myo-inositol cotransporter solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
| chr5_+_179125368 | 0.15 |
ENST00000502296.1 ENST00000504734.1 ENST00000415618.2 |
CANX |
calnexin |
| chr9_-_23821842 | 0.15 |
ENST00000544538.1 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr5_+_142149932 | 0.15 |
ENST00000274498.4 |
ARHGAP26 |
Rho GTPase activating protein 26 |
| chr14_+_57735614 | 0.15 |
ENST00000261558.3 |
AP5M1 |
adaptor-related protein complex 5, mu 1 subunit |
| chr19_-_19006890 | 0.14 |
ENST00000247005.6 |
GDF1 |
growth differentiation factor 1 |
| chr1_-_54304212 | 0.14 |
ENST00000540001.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr5_+_126112794 | 0.14 |
ENST00000261366.5 ENST00000395354.1 |
LMNB1 |
lamin B1 |
| chr3_+_14989076 | 0.14 |
ENST00000413118.1 ENST00000425241.1 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.4 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 0.7 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.3 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 0.3 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.1 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.3 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.1 | 0.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.2 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.4 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.4 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.2 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.5 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.5 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.6 | GO:0031228 | intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.4 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.0 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.2 | 0.9 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 0.5 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.5 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.8 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 1.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.1 | 1.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.2 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.1 | 0.4 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.1 | 0.8 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.6 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.1 | 0.4 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 1.0 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.3 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 0.2 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.2 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.2 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.0 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 1.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.3 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.3 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.3 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.1 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) |
| 0.0 | 0.2 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.0 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.3 | GO:0016505 | peptidase activator activity involved in apoptotic process(GO:0016505) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.1 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.7 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.4 | 1.6 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.3 | 0.9 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.3 | 0.6 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 | 0.6 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.2 | 0.5 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.1 | 0.6 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.4 | GO:0071789 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 0.4 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.1 | 0.7 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.1 | 0.4 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.1 | 0.7 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.4 | GO:1902661 | positive regulation of glucose mediated signaling pathway(GO:1902661) |
| 0.1 | 0.4 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.1 | 0.8 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.6 | GO:0032252 | secretory granule localization(GO:0032252) |
| 0.1 | 0.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.3 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.3 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.1 | 0.4 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 | 0.3 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.1 | 0.5 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.2 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.1 | 0.2 | GO:1900114 | positive regulation of histone H3-K9 trimethylation(GO:1900114) |
| 0.1 | 0.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.9 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.5 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.5 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.1 | 0.2 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.2 | GO:0060178 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.2 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.1 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 1.0 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.2 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.5 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.2 | GO:1903973 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.1 | 0.3 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.1 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.1 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.2 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.1 | 0.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.2 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.2 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.5 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.5 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.1 | GO:1902822 | cleavage furrow ingression(GO:0036090) regulation of late endosome to lysosome transport(GO:1902822) |
| 0.0 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.3 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.2 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.3 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.2 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.3 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.2 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.2 | GO:1901628 | positive regulation of postsynaptic membrane organization(GO:1901628) positive regulation of receptor clustering(GO:1903911) |
| 0.0 | 1.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:0036496 | regulation of translational initiation by eIF2 alpha dephosphorylation(GO:0036496) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0071072 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) negative regulation of phospholipid biosynthetic process(GO:0071072) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.3 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:2000987 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.5 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.3 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0061081 | positive regulation of macrophage cytokine production(GO:0060907) positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) |
| 0.0 | 0.1 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:0061074 | regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.0 | 0.1 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.0 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.2 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:0019856 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:1902965 | terminal web assembly(GO:1902896) regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.1 | GO:0010767 | regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage(GO:0010767) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0072364 | regulation of cellular ketone metabolic process by regulation of transcription from RNA polymerase II promoter(GO:0072364) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.2 | GO:0032341 | aldosterone metabolic process(GO:0032341) aldosterone biosynthetic process(GO:0032342) |
| 0.0 | 0.5 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.1 | GO:0032072 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.2 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.0 | 0.1 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.2 | GO:0015791 | polyol transport(GO:0015791) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.0 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) regulation of toll-like receptor 9 signaling pathway(GO:0034163) |
| 0.0 | 0.2 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.0 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.0 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.7 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.3 | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 0.1 | GO:0072396 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.1 | GO:0019086 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.0 | 0.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.0 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.2 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.0 | GO:0072054 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.0 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.0 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) negative regulation of gene silencing(GO:0060969) |
| 0.0 | 0.6 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0060897 | neural plate regionalization(GO:0060897) |
| 0.0 | 0.2 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:1904690 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.0 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.0 | 0.0 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.2 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.1 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.0 | GO:0042247 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.1 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.2 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.0 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.1 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.5 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 2.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.7 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.0 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.0 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.0 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |