Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
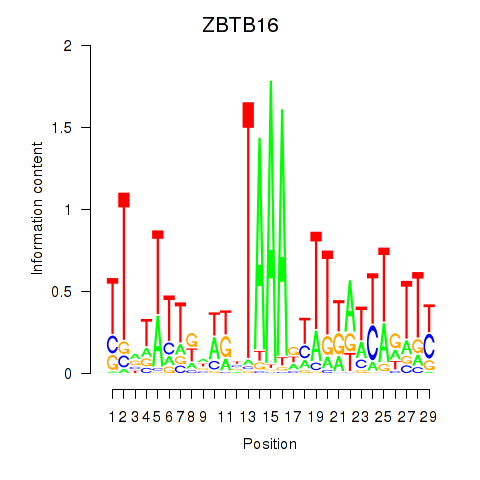
Results for ZBTB16
Z-value: 0.66
Transcription factors associated with ZBTB16
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB16
|
ENSG00000109906.9 | ZBTB16 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB16 | hg19_v2_chr11_+_113930291_113930339 | -0.21 | 4.3e-01 | Click! |
Activity profile of ZBTB16 motif
Sorted Z-values of ZBTB16 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB16
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_55562479 | 1.21 |
ENST00000564609.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr5_-_55412774 | 0.93 |
ENST00000434982.2 |
ANKRD55 |
ankyrin repeat domain 55 |
| chr10_+_62538089 | 0.92 |
ENST00000519078.2 ENST00000395284.3 ENST00000316629.4 |
CDK1 |
cyclin-dependent kinase 1 |
| chr11_-_5255861 | 0.87 |
ENST00000380299.3 |
HBD |
hemoglobin, delta |
| chr10_+_5005445 | 0.83 |
ENST00000380872.4 |
AKR1C1 |
aldo-keto reductase family 1, member C1 |
| chr6_+_26273144 | 0.81 |
ENST00000377733.2 |
HIST1H2BI |
histone cluster 1, H2bi |
| chr12_+_118454500 | 0.78 |
ENST00000537315.1 ENST00000229043.3 ENST00000484086.2 ENST00000420967.1 ENST00000454402.2 ENST00000392542.2 ENST00000535092.1 |
RFC5 |
replication factor C (activator 1) 5, 36.5kDa |
| chr15_-_55562582 | 0.76 |
ENST00000396307.2 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr5_+_7654057 | 0.75 |
ENST00000537121.1 |
ADCY2 |
adenylate cyclase 2 (brain) |
| chr1_-_246357029 | 0.73 |
ENST00000391836.2 |
SMYD3 |
SET and MYND domain containing 3 |
| chr1_+_198608146 | 0.71 |
ENST00000367376.2 ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
| chr5_-_34919094 | 0.70 |
ENST00000341754.4 |
RAD1 |
RAD1 homolog (S. pombe) |
| chr8_-_30585439 | 0.63 |
ENST00000221130.5 |
GSR |
glutathione reductase |
| chr3_-_93692781 | 0.63 |
ENST00000394236.3 |
PROS1 |
protein S (alpha) |
| chr10_+_5005598 | 0.53 |
ENST00000442997.1 |
AKR1C1 |
aldo-keto reductase family 1, member C1 |
| chr22_+_38203898 | 0.52 |
ENST00000323205.6 ENST00000248924.6 ENST00000445195.1 |
GCAT |
glycine C-acetyltransferase |
| chr21_+_40817749 | 0.51 |
ENST00000380637.3 ENST00000380634.1 ENST00000458295.1 ENST00000440288.2 ENST00000380631.1 |
SH3BGR |
SH3 domain binding glutamic acid-rich protein |
| chr17_-_46623441 | 0.50 |
ENST00000330070.4 |
HOXB2 |
homeobox B2 |
| chr2_-_11606275 | 0.44 |
ENST00000381525.3 ENST00000362009.4 |
E2F6 |
E2F transcription factor 6 |
| chr4_+_154622652 | 0.44 |
ENST00000260010.6 |
TLR2 |
toll-like receptor 2 |
| chr16_-_85722530 | 0.43 |
ENST00000253462.3 |
GINS2 |
GINS complex subunit 2 (Psf2 homolog) |
| chr4_+_74301880 | 0.43 |
ENST00000395792.2 ENST00000226359.2 |
AFP |
alpha-fetoprotein |
| chr4_-_99851766 | 0.42 |
ENST00000450253.2 |
EIF4E |
eukaryotic translation initiation factor 4E |
| chr16_+_20817761 | 0.41 |
ENST00000568046.1 ENST00000261377.6 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr16_-_20817753 | 0.41 |
ENST00000389345.5 ENST00000300005.3 ENST00000357967.4 ENST00000569729.1 |
ERI2 |
ERI1 exoribonuclease family member 2 |
| chr7_-_32534850 | 0.41 |
ENST00000409952.3 ENST00000409909.3 |
LSM5 |
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr11_-_72070206 | 0.41 |
ENST00000544382.1 |
CLPB |
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr8_-_33370607 | 0.39 |
ENST00000360742.5 ENST00000523305.1 |
TTI2 |
TELO2 interacting protein 2 |
| chr3_+_150126101 | 0.38 |
ENST00000361875.3 ENST00000361136.2 |
TSC22D2 |
TSC22 domain family, member 2 |
| chr1_-_44818599 | 0.37 |
ENST00000537474.1 |
ERI3 |
ERI1 exoribonuclease family member 3 |
| chr17_-_40134339 | 0.37 |
ENST00000587727.1 |
DNAJC7 |
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr4_-_76957214 | 0.37 |
ENST00000306621.3 |
CXCL11 |
chemokine (C-X-C motif) ligand 11 |
| chr7_+_120591170 | 0.36 |
ENST00000431467.1 |
ING3 |
inhibitor of growth family, member 3 |
| chr3_-_138312971 | 0.35 |
ENST00000485115.1 ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70 |
centrosomal protein 70kDa |
| chrX_+_65382381 | 0.35 |
ENST00000519389.1 |
HEPH |
hephaestin |
| chr10_+_5135981 | 0.35 |
ENST00000380554.3 |
AKR1C3 |
aldo-keto reductase family 1, member C3 |
| chr3_-_138313161 | 0.33 |
ENST00000489254.1 ENST00000474781.1 ENST00000462419.1 ENST00000264982.3 |
CEP70 |
centrosomal protein 70kDa |
| chr9_-_21351377 | 0.32 |
ENST00000380210.1 |
IFNA6 |
interferon, alpha 6 |
| chrX_-_23926004 | 0.31 |
ENST00000379226.4 ENST00000379220.3 |
APOO |
apolipoprotein O |
| chr12_+_53836339 | 0.30 |
ENST00000549135.1 |
PRR13 |
proline rich 13 |
| chr6_+_27925019 | 0.30 |
ENST00000244623.1 |
OR2B6 |
olfactory receptor, family 2, subfamily B, member 6 |
| chr21_-_35284635 | 0.30 |
ENST00000429238.1 |
AP000304.12 |
AP000304.12 |
| chr7_+_106505912 | 0.30 |
ENST00000359195.3 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr1_-_100231349 | 0.30 |
ENST00000287474.5 ENST00000414213.1 |
FRRS1 |
ferric-chelate reductase 1 |
| chr17_-_18266797 | 0.29 |
ENST00000316694.3 ENST00000539052.1 |
SHMT1 |
serine hydroxymethyltransferase 1 (soluble) |
| chr19_+_36142147 | 0.28 |
ENST00000590618.1 |
COX6B1 |
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr12_+_48166978 | 0.28 |
ENST00000442218.2 |
SLC48A1 |
solute carrier family 48 (heme transporter), member 1 |
| chr19_-_15087839 | 0.28 |
ENST00000600144.1 |
SLC1A6 |
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr16_-_18908196 | 0.28 |
ENST00000565324.1 ENST00000561947.1 |
SMG1 |
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr1_-_11159887 | 0.27 |
ENST00000544779.1 ENST00000304457.7 ENST00000376936.4 |
EXOSC10 |
exosome component 10 |
| chr12_-_113772835 | 0.27 |
ENST00000552014.1 ENST00000548186.1 ENST00000202831.3 ENST00000549181.1 |
SLC8B1 |
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr5_+_180682720 | 0.27 |
ENST00000599439.1 |
AC008443.1 |
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
| chr6_+_42749759 | 0.26 |
ENST00000314073.5 |
GLTSCR1L |
GLTSCR1-like |
| chrX_+_118602363 | 0.26 |
ENST00000317881.8 |
SLC25A5 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr9_-_15472730 | 0.26 |
ENST00000481862.1 |
PSIP1 |
PC4 and SFRS1 interacting protein 1 |
| chr4_-_99850243 | 0.26 |
ENST00000280892.6 ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E |
eukaryotic translation initiation factor 4E |
| chr15_+_75628419 | 0.26 |
ENST00000567377.1 ENST00000562789.1 ENST00000568301.1 |
COMMD4 |
COMM domain containing 4 |
| chr15_+_75628394 | 0.25 |
ENST00000564815.1 ENST00000338995.6 |
COMMD4 |
COMM domain containing 4 |
| chr15_+_75628232 | 0.24 |
ENST00000267935.8 ENST00000567195.1 |
COMMD4 |
COMM domain containing 4 |
| chr12_-_68696652 | 0.24 |
ENST00000539972.1 |
MDM1 |
Mdm1 nuclear protein homolog (mouse) |
| chr5_+_159848854 | 0.24 |
ENST00000517480.1 ENST00000520452.1 ENST00000393964.1 |
PTTG1 |
pituitary tumor-transforming 1 |
| chr8_-_8318847 | 0.23 |
ENST00000521218.1 |
CTA-398F10.2 |
CTA-398F10.2 |
| chr4_-_185275104 | 0.23 |
ENST00000317596.3 |
RP11-290F5.2 |
RP11-290F5.2 |
| chr6_+_74171301 | 0.22 |
ENST00000415954.2 ENST00000498286.1 ENST00000370305.1 ENST00000370300.4 |
MTO1 |
mitochondrial tRNA translation optimization 1 |
| chr1_+_15256230 | 0.22 |
ENST00000376028.4 ENST00000400798.2 |
KAZN |
kazrin, periplakin interacting protein |
| chr19_+_24269981 | 0.20 |
ENST00000339642.6 ENST00000357002.4 |
ZNF254 |
zinc finger protein 254 |
| chr19_-_23869999 | 0.20 |
ENST00000601935.1 ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675 |
zinc finger protein 675 |
| chr4_-_140223670 | 0.20 |
ENST00000394228.1 ENST00000539387.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr4_-_83769996 | 0.20 |
ENST00000511338.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr3_+_44840679 | 0.19 |
ENST00000425755.1 |
KIF15 |
kinesin family member 15 |
| chr6_-_49712123 | 0.19 |
ENST00000263045.4 |
CRISP3 |
cysteine-rich secretory protein 3 |
| chr3_-_137893721 | 0.19 |
ENST00000505015.2 ENST00000260803.4 |
DBR1 |
debranching RNA lariats 1 |
| chr5_+_159848807 | 0.19 |
ENST00000352433.5 |
PTTG1 |
pituitary tumor-transforming 1 |
| chr12_-_21487829 | 0.18 |
ENST00000445053.1 ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2 |
solute carrier organic anion transporter family, member 1A2 |
| chr10_-_47239738 | 0.18 |
ENST00000413193.2 |
AGAP10 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr6_+_136172820 | 0.17 |
ENST00000308191.6 |
PDE7B |
phosphodiesterase 7B |
| chr11_+_67351213 | 0.17 |
ENST00000398603.1 |
GSTP1 |
glutathione S-transferase pi 1 |
| chr1_+_241695670 | 0.17 |
ENST00000366557.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr8_+_104831472 | 0.17 |
ENST00000262231.10 ENST00000507740.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr17_+_32597232 | 0.17 |
ENST00000378569.2 ENST00000200307.4 ENST00000394627.1 ENST00000394630.3 |
CCL7 |
chemokine (C-C motif) ligand 7 |
| chr10_+_48189612 | 0.17 |
ENST00000453919.1 |
AGAP9 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 9 |
| chr9_-_21239978 | 0.17 |
ENST00000380222.2 |
IFNA14 |
interferon, alpha 14 |
| chr6_-_34639733 | 0.16 |
ENST00000374021.1 |
C6orf106 |
chromosome 6 open reading frame 106 |
| chr8_+_104831554 | 0.16 |
ENST00000408894.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr11_+_67351019 | 0.16 |
ENST00000398606.3 |
GSTP1 |
glutathione S-transferase pi 1 |
| chr19_+_21265028 | 0.16 |
ENST00000291770.7 |
ZNF714 |
zinc finger protein 714 |
| chr7_-_16872932 | 0.16 |
ENST00000419572.2 ENST00000412973.1 |
AGR2 |
anterior gradient 2 |
| chr2_+_131862872 | 0.16 |
ENST00000439822.2 |
PLEKHB2 |
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr3_-_155524049 | 0.15 |
ENST00000534941.1 ENST00000340171.2 |
C3orf33 |
chromosome 3 open reading frame 33 |
| chrX_+_83116142 | 0.15 |
ENST00000329312.4 |
CYLC1 |
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr4_-_140223614 | 0.15 |
ENST00000394223.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr7_-_104909435 | 0.15 |
ENST00000357311.3 |
SRPK2 |
SRSF protein kinase 2 |
| chr21_+_37692481 | 0.14 |
ENST00000400485.1 |
MORC3 |
MORC family CW-type zinc finger 3 |
| chr2_+_233527443 | 0.13 |
ENST00000410095.1 |
EFHD1 |
EF-hand domain family, member D1 |
| chr19_-_45004556 | 0.13 |
ENST00000587047.1 ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180 |
zinc finger protein 180 |
| chr19_+_13135386 | 0.13 |
ENST00000360105.4 ENST00000588228.1 ENST00000591028.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr2_-_73520667 | 0.12 |
ENST00000545030.1 ENST00000436467.2 |
EGR4 |
early growth response 4 |
| chr4_+_37962018 | 0.12 |
ENST00000504686.1 |
PTTG2 |
pituitary tumor-transforming 2 |
| chr4_-_90758227 | 0.11 |
ENST00000506691.1 ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chrX_+_71401526 | 0.11 |
ENST00000218432.5 ENST00000423432.2 ENST00000373669.2 |
PIN4 |
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr17_+_46908350 | 0.11 |
ENST00000258947.3 ENST00000509507.1 ENST00000448105.2 ENST00000570513.1 ENST00000509415.1 ENST00000513119.1 ENST00000416445.2 ENST00000508679.1 ENST00000505071.1 |
CALCOCO2 |
calcium binding and coiled-coil domain 2 |
| chr14_+_39703084 | 0.11 |
ENST00000553728.1 |
RP11-407N17.3 |
cTAGE family member 5 isoform 4 |
| chr6_-_49712147 | 0.11 |
ENST00000433368.2 ENST00000354620.4 |
CRISP3 |
cysteine-rich secretory protein 3 |
| chr1_+_32608566 | 0.11 |
ENST00000545542.1 |
KPNA6 |
karyopherin alpha 6 (importin alpha 7) |
| chr5_-_24645078 | 0.11 |
ENST00000264463.4 |
CDH10 |
cadherin 10, type 2 (T2-cadherin) |
| chr1_+_174769006 | 0.11 |
ENST00000489615.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr1_+_156095951 | 0.10 |
ENST00000448611.2 ENST00000368297.1 |
LMNA |
lamin A/C |
| chr12_-_27090896 | 0.10 |
ENST00000539625.1 ENST00000538727.1 |
ASUN |
asunder spermatogenesis regulator |
| chr2_-_242556900 | 0.10 |
ENST00000402545.1 ENST00000402136.1 |
THAP4 |
THAP domain containing 4 |
| chr2_+_131862900 | 0.10 |
ENST00000438882.2 ENST00000538982.1 ENST00000404460.1 |
PLEKHB2 |
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr4_-_90758118 | 0.10 |
ENST00000420646.2 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr20_+_31407692 | 0.10 |
ENST00000375571.5 |
MAPRE1 |
microtubule-associated protein, RP/EB family, member 1 |
| chr4_+_74606223 | 0.09 |
ENST00000307407.3 ENST00000401931.1 |
IL8 |
interleukin 8 |
| chr4_+_74347400 | 0.09 |
ENST00000226355.3 |
AFM |
afamin |
| chr3_-_146262488 | 0.09 |
ENST00000487389.1 |
PLSCR1 |
phospholipid scramblase 1 |
| chr5_+_140220769 | 0.09 |
ENST00000531613.1 ENST00000378123.3 |
PCDHA8 |
protocadherin alpha 8 |
| chr4_-_48082192 | 0.09 |
ENST00000507351.1 |
TXK |
TXK tyrosine kinase |
| chr5_+_173763250 | 0.08 |
ENST00000515513.1 ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1 |
RP11-267A15.1 |
| chr15_-_74658493 | 0.08 |
ENST00000419019.2 ENST00000569662.1 |
CYP11A1 |
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr5_+_96212185 | 0.07 |
ENST00000379904.4 |
ERAP2 |
endoplasmic reticulum aminopeptidase 2 |
| chr17_+_61086917 | 0.07 |
ENST00000424789.2 ENST00000389520.4 |
TANC2 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr15_-_85197501 | 0.07 |
ENST00000434634.2 |
WDR73 |
WD repeat domain 73 |
| chr2_+_89901292 | 0.07 |
ENST00000448155.2 |
IGKV1D-39 |
immunoglobulin kappa variable 1D-39 |
| chr1_+_178310581 | 0.06 |
ENST00000462775.1 |
RASAL2 |
RAS protein activator like 2 |
| chr19_+_45542295 | 0.06 |
ENST00000221455.3 ENST00000391953.4 ENST00000588936.1 |
CLASRP |
CLK4-associating serine/arginine rich protein |
| chr3_-_164796269 | 0.05 |
ENST00000264382.3 |
SI |
sucrase-isomaltase (alpha-glucosidase) |
| chr5_-_9630463 | 0.05 |
ENST00000382492.2 |
TAS2R1 |
taste receptor, type 2, member 1 |
| chr16_+_81070792 | 0.05 |
ENST00000564241.1 ENST00000565237.1 |
ATMIN |
ATM interactor |
| chrX_-_49089771 | 0.05 |
ENST00000376251.1 ENST00000323022.5 ENST00000376265.2 |
CACNA1F |
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr7_-_38289173 | 0.05 |
ENST00000436911.2 |
TRGC2 |
T cell receptor gamma constant 2 |
| chr4_+_106631966 | 0.05 |
ENST00000360505.5 ENST00000510865.1 ENST00000509336.1 |
GSTCD |
glutathione S-transferase, C-terminal domain containing |
| chr12_-_75905374 | 0.05 |
ENST00000438169.2 ENST00000229214.4 |
KRR1 |
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr15_-_74658519 | 0.05 |
ENST00000450547.1 ENST00000358632.4 |
CYP11A1 |
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr7_+_16828866 | 0.05 |
ENST00000597084.1 |
AC073333.1 |
Uncharacterized protein |
| chr1_+_76251879 | 0.04 |
ENST00000535300.1 ENST00000319942.3 |
RABGGTB |
Rab geranylgeranyltransferase, beta subunit |
| chrX_+_115301975 | 0.04 |
ENST00000371906.4 |
AGTR2 |
angiotensin II receptor, type 2 |
| chr1_-_13452656 | 0.04 |
ENST00000376132.3 |
PRAMEF13 |
PRAME family member 13 |
| chr2_+_102413726 | 0.04 |
ENST00000350878.4 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr17_-_37607497 | 0.04 |
ENST00000394287.3 ENST00000300651.6 |
MED1 |
mediator complex subunit 1 |
| chr2_-_89310012 | 0.04 |
ENST00000493819.1 |
IGKV1-9 |
immunoglobulin kappa variable 1-9 |
| chrX_+_140982452 | 0.04 |
ENST00000544766.1 |
MAGEC3 |
melanoma antigen family C, 3 |
| chr12_+_10460549 | 0.04 |
ENST00000543420.1 ENST00000543777.1 |
KLRD1 |
killer cell lectin-like receptor subfamily D, member 1 |
| chr16_+_19183671 | 0.04 |
ENST00000562711.2 |
SYT17 |
synaptotagmin XVII |
| chr3_+_46395219 | 0.04 |
ENST00000445132.2 ENST00000292301.4 |
CCR2 |
chemokine (C-C motif) receptor 2 |
| chr17_+_21729593 | 0.04 |
ENST00000581769.1 ENST00000584755.1 |
UBBP4 |
ubiquitin B pseudogene 4 |
| chr8_+_42552533 | 0.04 |
ENST00000289957.2 |
CHRNB3 |
cholinergic receptor, nicotinic, beta 3 (neuronal) |
| chr1_-_13673511 | 0.04 |
ENST00000344998.3 ENST00000334600.6 |
PRAMEF14 |
PRAME family member 14 |
| chr5_+_67586465 | 0.03 |
ENST00000336483.5 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr7_+_7606497 | 0.03 |
ENST00000340080.4 ENST00000405785.1 ENST00000433635.1 |
MIOS |
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr19_+_57640011 | 0.03 |
ENST00000598197.1 |
USP29 |
ubiquitin specific peptidase 29 |
| chr7_-_107770794 | 0.03 |
ENST00000205386.4 ENST00000418464.1 ENST00000388781.3 ENST00000388780.3 ENST00000414450.2 |
LAMB4 |
laminin, beta 4 |
| chr3_-_37216055 | 0.03 |
ENST00000336686.4 |
LRRFIP2 |
leucine rich repeat (in FLII) interacting protein 2 |
| chr4_+_71108300 | 0.03 |
ENST00000304954.3 |
CSN3 |
casein kappa |
| chr4_+_71263599 | 0.03 |
ENST00000399575.2 |
PROL1 |
proline rich, lacrimal 1 |
| chr13_+_76123883 | 0.03 |
ENST00000377595.3 |
UCHL3 |
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr15_-_50406680 | 0.03 |
ENST00000559829.1 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
| chr3_+_51663407 | 0.03 |
ENST00000432863.1 ENST00000296477.3 |
RAD54L2 |
RAD54-like 2 (S. cerevisiae) |
| chr2_+_204801471 | 0.03 |
ENST00000316386.6 ENST00000435193.1 |
ICOS |
inducible T-cell co-stimulator |
| chr3_+_40518599 | 0.02 |
ENST00000314686.5 ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619 |
zinc finger protein 619 |
| chr1_+_192605252 | 0.02 |
ENST00000391995.2 ENST00000543215.1 |
RGS13 |
regulator of G-protein signaling 13 |
| chr19_-_19932501 | 0.02 |
ENST00000540806.2 ENST00000590766.1 ENST00000587452.1 ENST00000545006.1 ENST00000590319.1 ENST00000587461.1 ENST00000450683.2 ENST00000443905.2 ENST00000590274.1 |
ZNF506 CTC-559E9.4 |
zinc finger protein 506 CTC-559E9.4 |
| chr16_+_89984287 | 0.02 |
ENST00000555147.1 |
MC1R |
melanocortin 1 receptor (alpha melanocyte stimulating hormone receptor) |
| chr4_+_88532028 | 0.02 |
ENST00000282478.7 |
DSPP |
dentin sialophosphoprotein |
| chr14_+_58754751 | 0.02 |
ENST00000598233.1 |
AL132989.1 |
AL132989.1 |
| chr1_+_12916941 | 0.02 |
ENST00000240189.2 |
PRAMEF2 |
PRAME family member 2 |
| chr11_+_120255997 | 0.02 |
ENST00000532993.1 |
ARHGEF12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr6_+_134758827 | 0.01 |
ENST00000431422.1 |
LINC01010 |
long intergenic non-protein coding RNA 1010 |
| chr3_+_160394940 | 0.01 |
ENST00000320767.2 |
ARL14 |
ADP-ribosylation factor-like 14 |
| chr2_+_1488435 | 0.01 |
ENST00000446278.1 ENST00000469607.1 |
TPO |
thyroid peroxidase |
| chrY_+_26997726 | 0.01 |
ENST00000382296.2 |
DAZ4 |
deleted in azoospermia 4 |
| chr1_+_12851545 | 0.01 |
ENST00000332296.7 |
PRAMEF1 |
PRAME family member 1 |
| chr18_-_64271363 | 0.01 |
ENST00000262150.2 |
CDH19 |
cadherin 19, type 2 |
| chr19_+_21264980 | 0.01 |
ENST00000596053.1 ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714 |
zinc finger protein 714 |
| chr4_+_96012614 | 0.01 |
ENST00000264568.4 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr19_-_29704448 | 0.01 |
ENST00000304863.4 |
UQCRFS1 |
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr3_-_120365866 | 0.01 |
ENST00000475447.2 |
HGD |
homogentisate 1,2-dioxygenase |
| chr12_+_21284118 | 0.00 |
ENST00000256958.2 |
SLCO1B1 |
solute carrier organic anion transporter family, member 1B1 |
| chr3_-_15382875 | 0.00 |
ENST00000408919.3 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
| chr9_+_105757590 | 0.00 |
ENST00000374798.3 ENST00000487798.1 |
CYLC2 |
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr3_-_146262637 | 0.00 |
ENST00000472349.1 ENST00000342435.4 |
PLSCR1 |
phospholipid scramblase 1 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.9 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 0.8 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.5 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.7 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.2 | 0.6 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 0.7 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 0.8 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.5 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.7 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 2.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.4 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.3 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 0.1 | 0.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.3 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.1 | 0.3 | GO:0070905 | serine binding(GO:0070905) |
| 0.1 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.9 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.3 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 1.0 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.2 | 1.7 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.2 | 0.5 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.2 | 0.7 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.2 | 0.5 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.2 | 0.8 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 0.9 | GO:0014038 | regulation of Schwann cell differentiation(GO:0014038) |
| 0.1 | 0.4 | GO:0032289 | central nervous system myelin formation(GO:0032289) detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.7 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) bone marrow development(GO:0048539) |
| 0.1 | 0.3 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.1 | 0.3 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.1 | 0.3 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 0.7 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 | 0.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.3 | GO:0070778 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.1 | 0.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.3 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) secretory granule localization(GO:0032252) |
| 0.0 | 0.3 | GO:1901029 | adenine transport(GO:0015853) negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.2 | GO:0051621 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.6 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.3 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.4 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 1.5 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.2 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.5 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.0 | GO:0002434 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) T cell extravasation(GO:0072683) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) negative regulation of eosinophil activation(GO:1902567) positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.6 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.5 | GO:2000816 | negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 0.7 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 2.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.4 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.8 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.3 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.1 | 0.4 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.2 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.0 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.7 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |