Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
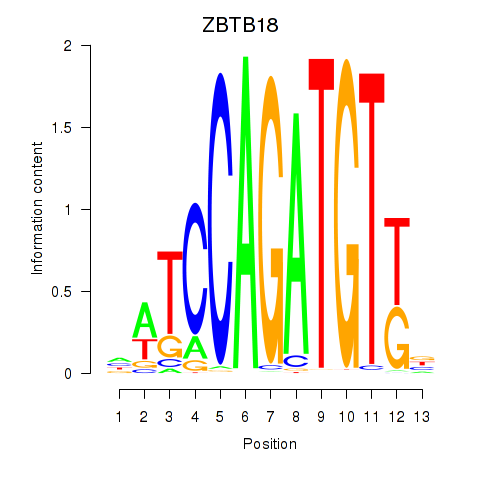
Results for ZBTB18
Z-value: 1.70
Transcription factors associated with ZBTB18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB18
|
ENSG00000179456.9 | ZBTB18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB18 | hg19_v2_chr1_+_244214577_244214593 | 0.37 | 1.6e-01 | Click! |
Activity profile of ZBTB18 motif
Sorted Z-values of ZBTB18 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB18
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_189839046 | 7.99 |
ENST00000304636.3 ENST00000317840.5 |
COL3A1 |
collagen, type III, alpha 1 |
| chr8_-_49834299 | 6.39 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr8_-_49833978 | 5.99 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr10_-_95241951 | 5.69 |
ENST00000358334.5 ENST00000359263.4 ENST00000371488.3 |
MYOF |
myoferlin |
| chr10_-_95242044 | 5.64 |
ENST00000371501.4 ENST00000371502.4 ENST00000371489.1 |
MYOF |
myoferlin |
| chr2_-_163099885 | 5.06 |
ENST00000443424.1 |
FAP |
fibroblast activation protein, alpha |
| chr2_-_163100045 | 4.86 |
ENST00000188790.4 |
FAP |
fibroblast activation protein, alpha |
| chr8_+_1772132 | 4.70 |
ENST00000349830.3 ENST00000520359.1 ENST00000518288.1 ENST00000398560.1 |
ARHGEF10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr1_-_153518270 | 4.62 |
ENST00000354332.4 ENST00000368716.4 |
S100A4 |
S100 calcium binding protein A4 |
| chrX_-_101771645 | 4.00 |
ENST00000289373.4 |
TMSB15A |
thymosin beta 15a |
| chr17_-_15168624 | 3.85 |
ENST00000312280.3 ENST00000494511.1 ENST00000580584.1 |
PMP22 |
peripheral myelin protein 22 |
| chr12_+_1738363 | 3.78 |
ENST00000397196.2 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
| chr12_-_104443890 | 3.73 |
ENST00000547583.1 ENST00000360814.4 ENST00000546851.1 |
GLT8D2 |
glycosyltransferase 8 domain containing 2 |
| chr21_+_30502806 | 3.49 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr1_-_156647189 | 3.32 |
ENST00000368223.3 |
NES |
nestin |
| chr7_+_100770328 | 2.84 |
ENST00000223095.4 ENST00000445463.2 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr2_+_33359646 | 2.78 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_33359687 | 2.77 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr9_+_125137565 | 2.74 |
ENST00000373698.5 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr3_+_159570722 | 2.71 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chrX_+_99899180 | 2.68 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr11_+_32112431 | 2.46 |
ENST00000054950.3 |
RCN1 |
reticulocalbin 1, EF-hand calcium binding domain |
| chr9_+_116263778 | 2.37 |
ENST00000394646.3 |
RGS3 |
regulator of G-protein signaling 3 |
| chr4_+_47487285 | 2.27 |
ENST00000273859.3 ENST00000504445.1 |
ATP10D |
ATPase, class V, type 10D |
| chr9_+_116263639 | 2.24 |
ENST00000343817.5 |
RGS3 |
regulator of G-protein signaling 3 |
| chr10_+_11206925 | 2.12 |
ENST00000354440.2 ENST00000315874.4 ENST00000427450.1 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr14_+_94577074 | 2.00 |
ENST00000444961.1 ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27 |
interferon, alpha-inducible protein 27 |
| chr7_+_90225796 | 1.94 |
ENST00000380050.3 |
CDK14 |
cyclin-dependent kinase 14 |
| chr5_+_49962495 | 1.91 |
ENST00000515175.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr9_+_97488939 | 1.88 |
ENST00000277198.2 ENST00000297979.5 |
C9orf3 |
chromosome 9 open reading frame 3 |
| chr4_-_111563076 | 1.80 |
ENST00000354925.2 ENST00000511990.1 |
PITX2 |
paired-like homeodomain 2 |
| chr12_-_56121612 | 1.72 |
ENST00000546939.1 |
CD63 |
CD63 molecule |
| chr12_-_56121580 | 1.72 |
ENST00000550776.1 |
CD63 |
CD63 molecule |
| chr9_-_13165457 | 1.70 |
ENST00000542239.1 ENST00000538841.1 ENST00000433359.2 |
MPDZ |
multiple PDZ domain protein |
| chr3_+_100211412 | 1.69 |
ENST00000323523.4 ENST00000403410.1 ENST00000449609.1 |
TMEM45A |
transmembrane protein 45A |
| chr2_-_145278475 | 1.63 |
ENST00000558170.2 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr17_+_32582293 | 1.57 |
ENST00000580907.1 ENST00000225831.4 |
CCL2 |
chemokine (C-C motif) ligand 2 |
| chr2_+_152214098 | 1.55 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr14_+_67707826 | 1.54 |
ENST00000261681.4 |
MPP5 |
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr15_+_67418047 | 1.42 |
ENST00000540846.2 |
SMAD3 |
SMAD family member 3 |
| chr12_-_56122220 | 1.35 |
ENST00000552692.1 |
CD63 |
CD63 molecule |
| chr16_+_6069586 | 1.35 |
ENST00000547372.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr1_+_38022572 | 1.25 |
ENST00000541606.1 |
DNALI1 |
dynein, axonemal, light intermediate chain 1 |
| chr12_-_56122124 | 1.23 |
ENST00000552754.1 |
CD63 |
CD63 molecule |
| chr13_-_33780133 | 1.20 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr12_-_56122426 | 1.16 |
ENST00000551173.1 |
CD63 |
CD63 molecule |
| chr2_-_152146385 | 1.15 |
ENST00000414946.1 ENST00000243346.5 |
NMI |
N-myc (and STAT) interactor |
| chr3_-_114035026 | 1.10 |
ENST00000570269.1 |
RP11-553L6.5 |
RP11-553L6.5 |
| chr4_+_75310851 | 1.09 |
ENST00000395748.3 ENST00000264487.2 |
AREG |
amphiregulin |
| chr4_+_75480629 | 1.08 |
ENST00000380846.3 |
AREGB |
amphiregulin B |
| chr4_+_75311019 | 0.98 |
ENST00000502307.1 |
AREG |
amphiregulin |
| chr3_-_37216055 | 0.98 |
ENST00000336686.4 |
LRRFIP2 |
leucine rich repeat (in FLII) interacting protein 2 |
| chr15_+_49715293 | 0.97 |
ENST00000267843.4 ENST00000560270.1 |
FGF7 |
fibroblast growth factor 7 |
| chr3_-_111314230 | 0.94 |
ENST00000317012.4 |
ZBED2 |
zinc finger, BED-type containing 2 |
| chr11_-_101454658 | 0.87 |
ENST00000344327.3 |
TRPC6 |
transient receptor potential cation channel, subfamily C, member 6 |
| chr19_+_11649532 | 0.82 |
ENST00000252456.2 ENST00000592923.1 ENST00000535659.2 |
CNN1 |
calponin 1, basic, smooth muscle |
| chr1_+_38022513 | 0.82 |
ENST00000296218.7 |
DNALI1 |
dynein, axonemal, light intermediate chain 1 |
| chr3_+_183894566 | 0.80 |
ENST00000439647.1 |
AP2M1 |
adaptor-related protein complex 2, mu 1 subunit |
| chr15_-_90358048 | 0.79 |
ENST00000300060.6 ENST00000560137.1 |
ANPEP |
alanyl (membrane) aminopeptidase |
| chr16_+_6069664 | 0.75 |
ENST00000422070.4 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr6_-_74363803 | 0.72 |
ENST00000355773.5 |
SLC17A5 |
solute carrier family 17 (acidic sugar transporter), member 5 |
| chr11_+_71900703 | 0.64 |
ENST00000393681.2 |
FOLR1 |
folate receptor 1 (adult) |
| chrX_-_153141302 | 0.62 |
ENST00000361699.4 ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM |
L1 cell adhesion molecule |
| chr5_+_140782351 | 0.57 |
ENST00000573521.1 |
PCDHGA9 |
protocadherin gamma subfamily A, 9 |
| chr4_-_100009856 | 0.56 |
ENST00000296412.8 |
ADH5 |
alcohol dehydrogenase 5 (class III), chi polypeptide |
| chr5_+_140792614 | 0.55 |
ENST00000398610.2 |
PCDHGA10 |
protocadherin gamma subfamily A, 10 |
| chr17_+_77020325 | 0.52 |
ENST00000311661.4 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
| chr16_-_18470696 | 0.48 |
ENST00000427999.2 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr17_+_77020224 | 0.47 |
ENST00000339142.2 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
| chr14_+_64970662 | 0.44 |
ENST00000556965.1 ENST00000554015.1 |
ZBTB1 |
zinc finger and BTB domain containing 1 |
| chr4_+_156824840 | 0.44 |
ENST00000536354.2 |
TDO2 |
tryptophan 2,3-dioxygenase |
| chr19_+_7584088 | 0.35 |
ENST00000394341.2 |
ZNF358 |
zinc finger protein 358 |
| chr7_+_120969045 | 0.35 |
ENST00000222462.2 |
WNT16 |
wingless-type MMTV integration site family, member 16 |
| chr8_+_24241789 | 0.35 |
ENST00000256412.4 ENST00000538205.1 |
ADAMDEC1 |
ADAM-like, decysin 1 |
| chr15_+_57210818 | 0.34 |
ENST00000438423.2 ENST00000267811.5 ENST00000452095.2 ENST00000559609.1 ENST00000333725.5 |
TCF12 |
transcription factor 12 |
| chr12_-_54121261 | 0.31 |
ENST00000549784.1 ENST00000262059.4 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
| chr15_+_57210961 | 0.31 |
ENST00000557843.1 |
TCF12 |
transcription factor 12 |
| chr17_+_32597232 | 0.31 |
ENST00000378569.2 ENST00000200307.4 ENST00000394627.1 ENST00000394630.3 |
CCL7 |
chemokine (C-C motif) ligand 7 |
| chr8_+_24241969 | 0.30 |
ENST00000522298.1 |
ADAMDEC1 |
ADAM-like, decysin 1 |
| chr15_+_55611128 | 0.30 |
ENST00000164305.5 ENST00000539642.1 |
PIGB |
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr3_-_52273098 | 0.30 |
ENST00000499914.2 ENST00000305533.5 ENST00000597542.1 |
TWF2 TLR9 |
twinfilin actin-binding protein 2 toll-like receptor 9 |
| chr12_-_53893227 | 0.29 |
ENST00000547488.1 |
MAP3K12 |
mitogen-activated protein kinase kinase kinase 12 |
| chr14_-_61748550 | 0.28 |
ENST00000555868.1 |
TMEM30B |
transmembrane protein 30B |
| chr3_+_121311966 | 0.28 |
ENST00000338040.4 |
FBXO40 |
F-box protein 40 |
| chr1_-_207206092 | 0.28 |
ENST00000359470.5 ENST00000461135.2 |
C1orf116 |
chromosome 1 open reading frame 116 |
| chr21_-_35883541 | 0.27 |
ENST00000399284.1 |
KCNE1 |
potassium voltage-gated channel, Isk-related family, member 1 |
| chr1_-_32210275 | 0.25 |
ENST00000440175.2 |
BAI2 |
brain-specific angiogenesis inhibitor 2 |
| chr9_+_118916082 | 0.25 |
ENST00000328252.3 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
| chr1_-_168698433 | 0.25 |
ENST00000367817.3 |
DPT |
dermatopontin |
| chr16_+_28914680 | 0.19 |
ENST00000564112.1 |
ATP2A1 |
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr18_+_6834472 | 0.18 |
ENST00000581099.1 ENST00000419673.2 ENST00000531294.1 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr12_+_112563335 | 0.18 |
ENST00000549358.1 ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1 |
TRAF-type zinc finger domain containing 1 |
| chr11_-_62323702 | 0.18 |
ENST00000530285.1 |
AHNAK |
AHNAK nucleoprotein |
| chr19_+_41119323 | 0.16 |
ENST00000599724.1 ENST00000597071.1 ENST00000243562.9 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr4_-_87855851 | 0.15 |
ENST00000473559.1 |
C4orf36 |
chromosome 4 open reading frame 36 |
| chr12_+_112563303 | 0.13 |
ENST00000412615.2 |
TRAFD1 |
TRAF-type zinc finger domain containing 1 |
| chr1_+_9005917 | 0.13 |
ENST00000549778.1 ENST00000480186.3 ENST00000377443.2 ENST00000377436.3 ENST00000377442.2 |
CA6 |
carbonic anhydrase VI |
| chr17_-_41174424 | 0.12 |
ENST00000355653.3 |
VAT1 |
vesicle amine transport 1 |
| chr12_-_53893399 | 0.11 |
ENST00000267079.2 |
MAP3K12 |
mitogen-activated protein kinase kinase kinase 12 |
| chr10_+_98064085 | 0.09 |
ENST00000419175.1 ENST00000371174.2 |
DNTT |
DNA nucleotidylexotransferase |
| chr19_-_15918936 | 0.07 |
ENST00000334920.2 |
OR10H1 |
olfactory receptor, family 10, subfamily H, member 1 |
| chr4_+_88571429 | 0.07 |
ENST00000339673.6 ENST00000282479.7 |
DMP1 |
dentin matrix acidic phosphoprotein 1 |
| chr10_+_7830125 | 0.07 |
ENST00000335698.4 ENST00000541227.1 |
ATP5C1 |
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr12_+_26126681 | 0.05 |
ENST00000542865.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chrX_+_153770421 | 0.03 |
ENST00000369609.5 ENST00000369607.1 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr16_-_49698136 | 0.02 |
ENST00000535559.1 |
ZNF423 |
zinc finger protein 423 |
| chr1_-_2458026 | 0.02 |
ENST00000435556.3 ENST00000378466.3 |
PANK4 |
pantothenate kinase 4 |
| chr22_-_37213554 | 0.02 |
ENST00000443735.1 |
PVALB |
parvalbumin |
| chr17_+_32612687 | 0.00 |
ENST00000305869.3 |
CCL11 |
chemokine (C-C motif) ligand 11 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 9.9 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.7 | 8.0 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.6 | 1.9 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.6 | 7.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.3 | 5.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 7.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.2 | 1.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.9 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 11.3 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 1.1 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 2.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 2.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 12.5 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 2.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 2.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.6 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 1.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 2.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 12.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 4.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 11.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 2.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 3.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 4.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.6 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 0.9 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.8 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 9.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 2.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 3.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 4.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 0.9 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 7.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 2.4 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.5 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 2.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.7 | 8.0 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.7 | 2.7 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.5 | 1.6 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.5 | 9.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.3 | 1.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.2 | 4.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 3.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 0.6 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.1 | 0.4 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 3.9 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 4.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 3.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 2.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.3 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 13.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 2.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 4.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 4.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.3 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 0.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 1.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.6 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 2.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.7 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 1.0 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.8 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 2.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 1.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 2.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 9.5 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 1.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 3.1 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 1.8 | GO:0019838 | growth factor binding(GO:0019838) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.4 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 2.5 | 9.9 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 1.0 | 8.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.9 | 2.8 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.9 | 11.3 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.6 | 5.5 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.6 | 1.8 | GO:0060578 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.6 | 7.2 | GO:2000680 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.5 | 1.6 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.4 | 1.6 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.4 | 1.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.3 | 2.1 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.3 | 1.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.3 | 3.8 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.3 | 1.4 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.3 | 2.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 3.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.2 | 1.0 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 | 1.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 0.6 | GO:0061713 | neural crest cell migration involved in heart formation(GO:0003147) cell migration involved in heart formation(GO:0060974) anterior neural tube closure(GO:0061713) |
| 0.2 | 6.2 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.2 | 0.6 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.2 | 2.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.4 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 3.3 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.1 | 0.4 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) |
| 0.1 | 4.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 0.4 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.4 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 2.3 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 0.3 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) positive regulation of interferon-alpha biosynthetic process(GO:0045356) negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.1 | 0.2 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.1 | 0.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 2.1 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 3.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 2.0 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.8 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.8 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.9 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 3.0 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.0 | 1.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 4.2 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.3 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.0 | 0.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.7 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |