Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
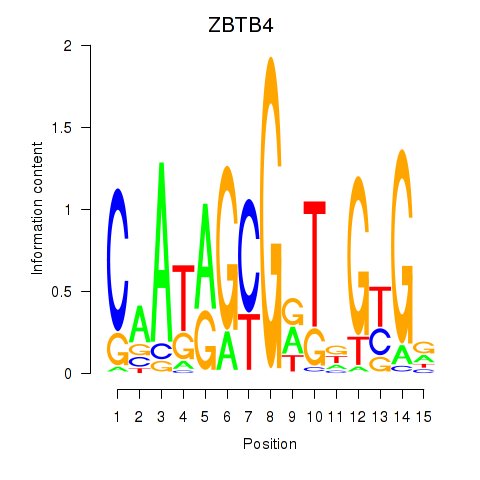
Results for ZBTB4
Z-value: 1.31
Transcription factors associated with ZBTB4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB4
|
ENSG00000174282.7 | ZBTB4 |
Activity profile of ZBTB4 motif
Sorted Z-values of ZBTB4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_86511569 | 4.79 |
ENST00000441050.1 |
PRSS23 |
protease, serine, 23 |
| chr14_-_69445968 | 4.37 |
ENST00000438964.2 |
ACTN1 |
actinin, alpha 1 |
| chr2_+_192110199 | 4.02 |
ENST00000304164.4 |
MYO1B |
myosin IB |
| chr2_+_192109911 | 3.97 |
ENST00000418908.1 ENST00000339514.4 ENST00000392318.3 |
MYO1B |
myosin IB |
| chr14_-_69445793 | 3.96 |
ENST00000538545.2 ENST00000394419.4 |
ACTN1 |
actinin, alpha 1 |
| chr8_+_94929077 | 3.72 |
ENST00000297598.4 ENST00000520614.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr14_-_69446034 | 3.31 |
ENST00000193403.6 |
ACTN1 |
actinin, alpha 1 |
| chr14_+_85996471 | 2.72 |
ENST00000330753.4 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr19_+_39138320 | 2.48 |
ENST00000424234.2 ENST00000390009.3 ENST00000589528.1 |
ACTN4 |
actinin, alpha 4 |
| chr8_+_94929168 | 2.39 |
ENST00000518107.1 ENST00000396200.3 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr1_-_154946825 | 2.33 |
ENST00000368453.4 ENST00000368450.1 ENST00000366442.2 |
SHC1 |
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr8_+_94929110 | 2.31 |
ENST00000520728.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr5_-_146889619 | 2.07 |
ENST00000343218.5 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr17_-_15165854 | 1.76 |
ENST00000395936.1 ENST00000395938.2 |
PMP22 |
peripheral myelin protein 22 |
| chr14_+_85996507 | 1.70 |
ENST00000554746.1 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr1_-_108742957 | 1.69 |
ENST00000565488.1 |
SLC25A24 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr16_-_89556942 | 1.55 |
ENST00000301030.4 |
ANKRD11 |
ankyrin repeat domain 11 |
| chrX_-_68385354 | 1.48 |
ENST00000361478.1 |
PJA1 |
praja ring finger 1, E3 ubiquitin protein ligase |
| chrX_-_68385274 | 1.39 |
ENST00000374584.3 ENST00000590146.1 |
PJA1 |
praja ring finger 1, E3 ubiquitin protein ligase |
| chr4_+_4388805 | 1.38 |
ENST00000504171.1 |
NSG1 |
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr15_+_25200074 | 1.36 |
ENST00000390687.4 ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr10_+_123922941 | 1.33 |
ENST00000360561.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr4_-_156298028 | 1.32 |
ENST00000433024.1 ENST00000379248.2 |
MAP9 |
microtubule-associated protein 9 |
| chr3_-_57583052 | 1.28 |
ENST00000496292.1 ENST00000489843.1 |
ARF4 |
ADP-ribosylation factor 4 |
| chr3_-_57583130 | 1.28 |
ENST00000303436.6 |
ARF4 |
ADP-ribosylation factor 4 |
| chr11_+_62623512 | 1.24 |
ENST00000377892.1 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr16_-_79634595 | 1.24 |
ENST00000326043.4 ENST00000393350.1 |
MAF |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog |
| chr15_+_25200108 | 1.23 |
ENST00000577949.1 ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF SNRPN |
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr11_+_62623544 | 1.19 |
ENST00000377890.2 ENST00000377891.2 ENST00000377889.2 |
SLC3A2 |
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr19_+_39138271 | 1.17 |
ENST00000252699.2 |
ACTN4 |
actinin, alpha 4 |
| chr3_-_57583185 | 1.14 |
ENST00000463880.1 |
ARF4 |
ADP-ribosylation factor 4 |
| chr4_-_156297949 | 1.13 |
ENST00000515654.1 |
MAP9 |
microtubule-associated protein 9 |
| chr3_+_148583043 | 1.11 |
ENST00000296046.3 |
CPA3 |
carboxypeptidase A3 (mast cell) |
| chr20_-_36793774 | 1.08 |
ENST00000361475.2 |
TGM2 |
transglutaminase 2 |
| chr4_-_156298087 | 1.03 |
ENST00000311277.4 |
MAP9 |
microtubule-associated protein 9 |
| chr8_-_29208183 | 1.01 |
ENST00000240100.2 |
DUSP4 |
dual specificity phosphatase 4 |
| chr11_-_72385437 | 0.99 |
ENST00000418754.2 ENST00000542969.2 ENST00000334456.5 |
PDE2A |
phosphodiesterase 2A, cGMP-stimulated |
| chr19_+_36631867 | 0.98 |
ENST00000588780.1 |
CAPNS1 |
calpain, small subunit 1 |
| chr11_+_65082289 | 0.94 |
ENST00000279249.2 |
CDC42EP2 |
CDC42 effector protein (Rho GTPase binding) 2 |
| chr17_-_9479128 | 0.92 |
ENST00000574431.1 |
STX8 |
syntaxin 8 |
| chr5_-_150948414 | 0.89 |
ENST00000261800.5 |
FAT2 |
FAT atypical cadherin 2 |
| chr9_-_131709858 | 0.89 |
ENST00000372586.3 |
DOLK |
dolichol kinase |
| chr17_-_5389477 | 0.88 |
ENST00000572834.1 ENST00000570848.1 ENST00000571971.1 ENST00000158771.4 |
DERL2 |
derlin 2 |
| chr8_-_27469196 | 0.81 |
ENST00000546343.1 ENST00000560566.1 |
CLU |
clusterin |
| chr13_-_33780133 | 0.78 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr14_+_74416989 | 0.76 |
ENST00000334571.2 ENST00000554920.1 |
COQ6 |
coenzyme Q6 monooxygenase |
| chr1_+_169075554 | 0.74 |
ENST00000367815.4 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr8_-_27468842 | 0.74 |
ENST00000523500.1 |
CLU |
clusterin |
| chr8_-_27468945 | 0.72 |
ENST00000405140.3 |
CLU |
clusterin |
| chr1_+_65886262 | 0.71 |
ENST00000371065.4 |
LEPROT |
leptin receptor overlapping transcript |
| chr22_+_50312379 | 0.71 |
ENST00000407217.3 ENST00000403427.3 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
| chr11_-_64546202 | 0.70 |
ENST00000377390.3 ENST00000227503.9 ENST00000377394.3 ENST00000422298.2 ENST00000334944.5 |
SF1 |
splicing factor 1 |
| chr2_+_182756701 | 0.67 |
ENST00000409001.1 |
SSFA2 |
sperm specific antigen 2 |
| chr22_+_50312316 | 0.67 |
ENST00000328268.4 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
| chr15_-_77363513 | 0.62 |
ENST00000267970.4 |
TSPAN3 |
tetraspanin 3 |
| chr8_-_74205851 | 0.62 |
ENST00000396467.1 |
RPL7 |
ribosomal protein L7 |
| chr1_+_196788887 | 0.61 |
ENST00000320493.5 ENST00000367424.4 ENST00000367421.3 |
CFHR1 CFHR2 |
complement factor H-related 1 complement factor H-related 2 |
| chr19_-_10530784 | 0.61 |
ENST00000593124.1 |
CDC37 |
cell division cycle 37 |
| chr10_-_101380121 | 0.57 |
ENST00000370495.4 |
SLC25A28 |
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr17_-_19265855 | 0.55 |
ENST00000440841.1 ENST00000395615.1 ENST00000461069.2 |
B9D1 |
B9 protein domain 1 |
| chr10_+_121485588 | 0.52 |
ENST00000361976.2 ENST00000369083.3 |
INPP5F |
inositol polyphosphate-5-phosphatase F |
| chr19_+_17622415 | 0.50 |
ENST00000252603.2 ENST00000600923.1 |
PGLS |
6-phosphogluconolactonase |
| chr17_-_48943706 | 0.50 |
ENST00000499247.2 |
TOB1 |
transducer of ERBB2, 1 |
| chr14_-_23822080 | 0.48 |
ENST00000397267.1 ENST00000354772.3 |
SLC22A17 |
solute carrier family 22, member 17 |
| chr19_-_39881669 | 0.47 |
ENST00000221266.7 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr3_-_119813264 | 0.47 |
ENST00000264235.8 |
GSK3B |
glycogen synthase kinase 3 beta |
| chr15_-_40212363 | 0.46 |
ENST00000299092.3 |
GPR176 |
G protein-coupled receptor 176 |
| chr12_-_46766577 | 0.46 |
ENST00000256689.5 |
SLC38A2 |
solute carrier family 38, member 2 |
| chr14_-_24701539 | 0.46 |
ENST00000534348.1 ENST00000524927.1 ENST00000250495.5 |
NEDD8-MDP1 NEDD8 |
NEDD8-MDP1 readthrough neural precursor cell expressed, developmentally down-regulated 8 |
| chr14_+_23305760 | 0.45 |
ENST00000311852.6 |
MMP14 |
matrix metallopeptidase 14 (membrane-inserted) |
| chr19_+_21106081 | 0.45 |
ENST00000300540.3 ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85 |
zinc finger protein 85 |
| chr15_-_40213080 | 0.44 |
ENST00000561100.1 |
GPR176 |
G protein-coupled receptor 176 |
| chr1_-_36615051 | 0.43 |
ENST00000373163.1 |
TRAPPC3 |
trafficking protein particle complex 3 |
| chr13_-_24007815 | 0.42 |
ENST00000382298.3 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr14_+_74417192 | 0.42 |
ENST00000554320.1 |
COQ6 |
coenzyme Q6 monooxygenase |
| chr10_-_53459319 | 0.41 |
ENST00000331173.4 |
CSTF2T |
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant |
| chr13_-_50018140 | 0.41 |
ENST00000410043.1 ENST00000347776.5 |
CAB39L |
calcium binding protein 39-like |
| chr6_-_49430886 | 0.40 |
ENST00000274813.3 |
MUT |
methylmalonyl CoA mutase |
| chr14_-_35099315 | 0.37 |
ENST00000396526.3 ENST00000396534.3 ENST00000355110.5 ENST00000557265.1 |
SNX6 |
sorting nexin 6 |
| chrX_+_153237740 | 0.37 |
ENST00000369982.4 |
TMEM187 |
transmembrane protein 187 |
| chr19_-_19144243 | 0.37 |
ENST00000594445.1 ENST00000452918.2 ENST00000600377.1 ENST00000337018.6 |
SUGP2 |
SURP and G patch domain containing 2 |
| chr1_-_36615065 | 0.36 |
ENST00000373166.3 ENST00000373159.1 ENST00000373162.1 |
TRAPPC3 |
trafficking protein particle complex 3 |
| chr19_+_16435625 | 0.36 |
ENST00000248071.5 ENST00000592003.1 |
KLF2 |
Kruppel-like factor 2 |
| chr17_+_45727204 | 0.36 |
ENST00000290158.4 |
KPNB1 |
karyopherin (importin) beta 1 |
| chr15_+_42697065 | 0.35 |
ENST00000565559.1 |
CAPN3 |
calpain 3, (p94) |
| chr10_-_35104185 | 0.35 |
ENST00000374789.3 ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3 |
par-3 family cell polarity regulator |
| chr1_+_203764742 | 0.34 |
ENST00000432282.1 ENST00000453771.1 ENST00000367214.1 ENST00000367212.3 ENST00000332127.4 |
ZC3H11A |
zinc finger CCCH-type containing 11A |
| chr20_-_36793663 | 0.33 |
ENST00000536701.1 ENST00000536724.1 |
TGM2 |
transglutaminase 2 |
| chr17_-_4046257 | 0.30 |
ENST00000381638.2 |
ZZEF1 |
zinc finger, ZZ-type with EF-hand domain 1 |
| chr2_+_207024306 | 0.30 |
ENST00000236957.5 ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2 |
eukaryotic translation elongation factor 1 beta 2 |
| chr1_+_210502238 | 0.30 |
ENST00000545154.1 ENST00000537898.1 ENST00000391905.3 ENST00000545781.1 ENST00000261458.3 ENST00000308852.6 |
HHAT |
hedgehog acyltransferase |
| chr1_-_203055129 | 0.29 |
ENST00000241651.4 |
MYOG |
myogenin (myogenic factor 4) |
| chr19_+_19144384 | 0.27 |
ENST00000392335.2 ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6 |
armadillo repeat containing 6 |
| chr1_+_196912902 | 0.27 |
ENST00000476712.2 ENST00000367415.5 |
CFHR2 |
complement factor H-related 2 |
| chr10_-_32345305 | 0.27 |
ENST00000302418.4 |
KIF5B |
kinesin family member 5B |
| chr1_+_43148059 | 0.27 |
ENST00000321358.7 ENST00000332220.6 |
YBX1 |
Y box binding protein 1 |
| chr12_+_57388230 | 0.27 |
ENST00000300098.1 |
GPR182 |
G protein-coupled receptor 182 |
| chr16_+_640201 | 0.26 |
ENST00000563109.1 |
RAB40C |
RAB40C, member RAS oncogene family |
| chr1_+_151254738 | 0.26 |
ENST00000336715.6 ENST00000324048.5 ENST00000368879.2 |
ZNF687 |
zinc finger protein 687 |
| chr1_+_2160134 | 0.25 |
ENST00000378536.4 |
SKI |
v-ski avian sarcoma viral oncogene homolog |
| chr1_-_165738085 | 0.24 |
ENST00000464650.1 ENST00000392129.6 |
TMCO1 |
transmembrane and coiled-coil domains 1 |
| chr14_-_45603657 | 0.23 |
ENST00000396062.3 |
FKBP3 |
FK506 binding protein 3, 25kDa |
| chr1_+_154947126 | 0.22 |
ENST00000368439.1 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
| chr19_-_39881777 | 0.22 |
ENST00000595564.1 ENST00000221265.3 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr1_+_154947148 | 0.21 |
ENST00000368436.1 ENST00000308987.5 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
| chr2_-_10952922 | 0.21 |
ENST00000272227.3 |
PDIA6 |
protein disulfide isomerase family A, member 6 |
| chr6_+_41606176 | 0.21 |
ENST00000441667.1 ENST00000230321.6 ENST00000373050.4 ENST00000446650.1 ENST00000435476.1 |
MDFI |
MyoD family inhibitor |
| chr5_-_115177247 | 0.21 |
ENST00000500945.2 |
ATG12 |
autophagy related 12 |
| chr10_+_105036909 | 0.20 |
ENST00000369849.4 |
INA |
internexin neuronal intermediate filament protein, alpha |
| chr22_-_29784519 | 0.18 |
ENST00000357586.2 ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1 |
adaptor-related protein complex 1, beta 1 subunit |
| chr11_-_64815333 | 0.18 |
ENST00000533842.1 ENST00000532802.1 ENST00000530139.1 ENST00000526516.1 |
NAALADL1 |
N-acetylated alpha-linked acidic dipeptidase-like 1 |
| chr19_+_36024310 | 0.16 |
ENST00000222286.4 |
GAPDHS |
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr4_+_71554196 | 0.16 |
ENST00000254803.2 |
UTP3 |
UTP3, small subunit (SSU) processome component, homolog (S. cerevisiae) |
| chr22_+_32439019 | 0.15 |
ENST00000266088.4 |
SLC5A1 |
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr2_-_241497374 | 0.15 |
ENST00000373318.2 ENST00000406958.1 ENST00000391987.1 ENST00000373320.4 |
ANKMY1 |
ankyrin repeat and MYND domain containing 1 |
| chr9_-_88896977 | 0.15 |
ENST00000311534.6 |
ISCA1 |
iron-sulfur cluster assembly 1 |
| chr14_-_74551096 | 0.15 |
ENST00000350259.4 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr13_+_28712614 | 0.15 |
ENST00000380958.3 |
PAN3 |
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr19_+_10531150 | 0.15 |
ENST00000352831.6 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr19_+_10828795 | 0.14 |
ENST00000389253.4 ENST00000355667.6 ENST00000408974.4 |
DNM2 |
dynamin 2 |
| chr19_-_18902106 | 0.14 |
ENST00000542601.2 ENST00000425807.1 ENST00000222271.2 |
COMP |
cartilage oligomeric matrix protein |
| chr10_+_76586348 | 0.13 |
ENST00000372724.1 ENST00000287239.4 ENST00000372714.1 |
KAT6B |
K(lysine) acetyltransferase 6B |
| chr2_+_155554797 | 0.12 |
ENST00000295101.2 |
KCNJ3 |
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr5_-_137090028 | 0.12 |
ENST00000314940.4 |
HNRNPA0 |
heterogeneous nuclear ribonucleoprotein A0 |
| chr19_-_9006766 | 0.11 |
ENST00000599436.1 |
MUC16 |
mucin 16, cell surface associated |
| chr19_+_17830051 | 0.11 |
ENST00000594625.1 ENST00000324096.4 ENST00000600186.1 ENST00000597735.1 |
MAP1S |
microtubule-associated protein 1S |
| chr3_+_40518599 | 0.11 |
ENST00000314686.5 ENST00000447116.2 ENST00000429348.2 ENST00000456778.1 |
ZNF619 |
zinc finger protein 619 |
| chr15_+_42696954 | 0.10 |
ENST00000337571.4 ENST00000569136.1 |
CAPN3 |
calpain 3, (p94) |
| chr11_-_61735029 | 0.10 |
ENST00000526640.1 |
FTH1 |
ferritin, heavy polypeptide 1 |
| chr14_+_24701819 | 0.09 |
ENST00000560139.1 ENST00000559910.1 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr9_-_88897426 | 0.09 |
ENST00000375991.4 ENST00000326094.4 |
ISCA1 |
iron-sulfur cluster assembly 1 |
| chr17_+_7487146 | 0.07 |
ENST00000396501.4 ENST00000584378.1 ENST00000423172.2 ENST00000579445.1 ENST00000585217.1 ENST00000581380.1 |
MPDU1 |
mannose-P-dolichol utilization defect 1 |
| chr19_+_47104493 | 0.07 |
ENST00000291295.9 ENST00000597743.1 |
CALM3 |
calmodulin 3 (phosphorylase kinase, delta) |
| chr11_-_46940074 | 0.07 |
ENST00000378623.1 ENST00000534404.1 |
LRP4 |
low density lipoprotein receptor-related protein 4 |
| chr2_-_241497390 | 0.07 |
ENST00000272972.3 ENST00000401804.1 ENST00000361678.4 ENST00000405523.3 |
ANKMY1 |
ankyrin repeat and MYND domain containing 1 |
| chr1_+_200842083 | 0.06 |
ENST00000304244.2 |
GPR25 |
G protein-coupled receptor 25 |
| chr2_+_70314579 | 0.06 |
ENST00000303577.5 |
PCBP1 |
poly(rC) binding protein 1 |
| chr19_+_10828724 | 0.06 |
ENST00000585892.1 ENST00000314646.5 ENST00000359692.6 |
DNM2 |
dynamin 2 |
| chr19_+_47759716 | 0.05 |
ENST00000221922.6 |
CCDC9 |
coiled-coil domain containing 9 |
| chr16_+_640055 | 0.05 |
ENST00000568586.1 ENST00000538492.1 ENST00000248139.3 |
RAB40C |
RAB40C, member RAS oncogene family |
| chr2_-_207024233 | 0.05 |
ENST00000423725.1 ENST00000233190.6 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr12_+_5541267 | 0.05 |
ENST00000423158.3 |
NTF3 |
neurotrophin 3 |
| chr1_+_43148625 | 0.05 |
ENST00000436427.1 |
YBX1 |
Y box binding protein 1 |
| chr2_+_152266392 | 0.03 |
ENST00000444746.2 ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1 |
RAP1 interacting factor homolog (yeast) |
| chr3_+_19988566 | 0.03 |
ENST00000273047.4 |
RAB5A |
RAB5A, member RAS oncogene family |
| chr9_-_71629010 | 0.03 |
ENST00000377276.2 |
PRKACG |
protein kinase, cAMP-dependent, catalytic, gamma |
| chr1_+_110754094 | 0.03 |
ENST00000369787.3 ENST00000413138.3 ENST00000438661.2 |
KCNC4 |
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr11_+_118754475 | 0.03 |
ENST00000292174.4 |
CXCR5 |
chemokine (C-X-C motif) receptor 5 |
| chr12_+_6561190 | 0.03 |
ENST00000544021.1 ENST00000266556.7 |
TAPBPL |
TAP binding protein-like |
| chr15_+_42697018 | 0.02 |
ENST00000397204.4 |
CAPN3 |
calpain 3, (p94) |
| chr10_+_17851362 | 0.02 |
ENST00000331429.2 ENST00000457317.1 |
MRC1L1 |
cDNA FLJ56855, highly similar to Macrophage mannose receptor 1 |
| chr17_-_79827808 | 0.02 |
ENST00000580685.1 |
ARHGDIA |
Rho GDP dissociation inhibitor (GDI) alpha |
| chr18_-_55253871 | 0.01 |
ENST00000382873.3 |
FECH |
ferrochelatase |
| chr14_+_24702099 | 0.01 |
ENST00000420554.2 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr15_-_75660919 | 0.01 |
ENST00000569482.1 ENST00000565683.1 ENST00000561615.1 ENST00000563622.1 ENST00000568374.1 ENST00000566256.1 ENST00000267978.5 |
MAN2C1 |
mannosidase, alpha, class 2C, member 1 |
| chr8_+_11660120 | 0.01 |
ENST00000220584.4 |
FDFT1 |
farnesyl-diphosphate farnesyltransferase 1 |
| chr12_-_80084594 | 0.01 |
ENST00000548426.1 |
PAWR |
PRKC, apoptosis, WT1, regulator |
| chr21_-_38445470 | 0.00 |
ENST00000399098.1 |
PIGP |
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr19_+_41770349 | 0.00 |
ENST00000602130.1 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr2_-_10952832 | 0.00 |
ENST00000540494.1 |
PDIA6 |
protein disulfide isomerase family A, member 6 |
| chr14_+_24702073 | 0.00 |
ENST00000399440.2 |
GMPR2 |
guanosine monophosphate reductase 2 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 2.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 3.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 3.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 2.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 8.4 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.5 | 11.6 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 8.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 1.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.3 | 2.3 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.3 | 4.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 2.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.2 | 2.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 1.7 | GO:0005347 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleotide transmembrane transporter activity(GO:0015216) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 1.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.2 | 0.5 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.2 | 1.0 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.2 | 1.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 3.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.9 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 2.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.5 | GO:0052833 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.9 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 3.6 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 0.3 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.1 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.5 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 1.2 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 1.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 6.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.0 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.0 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 2.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 3.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.4 | 1.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.3 | 1.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 1.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 1.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.6 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.7 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 0.4 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.4 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.2 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.8 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 1.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 0.8 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 1.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.5 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 2.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.5 | 15.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 2.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 3.1 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.0 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.9 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 1.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 2.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.7 | 4.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.7 | 11.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.6 | 9.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.5 | 2.4 | GO:0060356 | leucine import(GO:0060356) |
| 0.5 | 1.4 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.4 | 3.5 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.4 | 2.3 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.3 | 3.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) apical protein localization(GO:0045176) |
| 0.3 | 1.0 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.2 | 0.7 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.2 | 1.7 | GO:0015866 | ADP transport(GO:0015866) |
| 0.2 | 0.6 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.2 | 0.7 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 2.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 0.5 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.2 | 0.5 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.2 | 0.8 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.7 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.4 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.9 | GO:0046465 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.1 | 0.9 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 1.4 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.1 | 0.4 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.1 | 0.5 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.3 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 0.4 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 0.4 | GO:1990834 | response to odorant(GO:1990834) |
| 0.1 | 1.8 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.5 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 0.5 | GO:0032328 | alanine transport(GO:0032328) |
| 0.1 | 0.7 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.9 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 2.7 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 1.1 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.1 | 0.9 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.2 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.1 | 0.3 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 7.5 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 1.2 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.3 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.1 | 0.5 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.1 | 0.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 1.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.5 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.6 | GO:0098779 | mitophagy in response to mitochondrial depolarization(GO:0098779) |
| 0.0 | 0.4 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 1.0 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.7 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.3 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.3 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.9 | GO:0048512 | circadian behavior(GO:0048512) |
| 0.0 | 0.1 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.4 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.3 | 1.3 | GO:0097476 | NMDA glutamate receptor clustering(GO:0097114) spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.2 | 1.0 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.2 | 1.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.8 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.3 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.4 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.7 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 | 0.7 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.2 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) regulation of lactation(GO:1903487) |
| 0.1 | 0.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.6 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.7 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.3 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 0.3 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.6 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.3 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 1.2 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.6 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.5 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.3 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.0 | 1.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.8 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 11.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.4 | 3.5 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.4 | 2.3 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.2 | 1.4 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.2 | 2.6 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 3.6 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 2.3 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.9 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 8.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.4 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 0.7 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.8 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.2 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 2.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.0 | 0.1 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 3.7 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.9 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 2.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 4.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 1.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 3.7 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |