Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for ZBTB49
Z-value: 0.92
Transcription factors associated with ZBTB49
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB49
|
ENSG00000168826.11 | ZBTB49 |
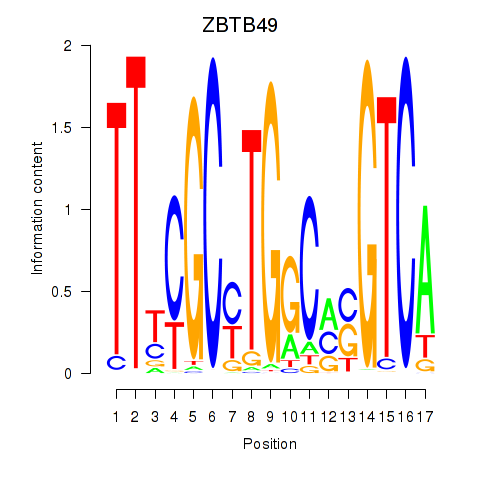
Activity profile of ZBTB49 motif
Sorted Z-values of ZBTB49 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB49
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_49604545 | 4.77 |
ENST00000371175.4 ENST00000229810.7 |
RHAG |
Rh-associated glycoprotein |
| chrX_-_151903184 | 4.18 |
ENST00000357916.4 ENST00000393869.3 |
MAGEA12 |
melanoma antigen family A, 12 |
| chrX_-_151903101 | 3.47 |
ENST00000393900.3 |
MAGEA12 |
melanoma antigen family A, 12 |
| chrX_+_30265256 | 2.93 |
ENST00000397548.2 |
MAGEB1 |
melanoma antigen family B, 1 |
| chrX_+_151903253 | 2.43 |
ENST00000452779.2 ENST00000370291.2 |
CSAG1 |
chondrosarcoma associated gene 1 |
| chrX_+_52780318 | 1.90 |
ENST00000375515.3 ENST00000276049.6 |
SSX2B |
synovial sarcoma, X breakpoint 2B |
| chr22_+_21996549 | 1.86 |
ENST00000248958.4 |
SDF2L1 |
stromal cell-derived factor 2-like 1 |
| chrX_+_9431324 | 1.74 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr7_-_103629963 | 1.34 |
ENST00000428762.1 ENST00000343529.5 ENST00000424685.2 |
RELN |
reelin |
| chr1_+_182808474 | 1.05 |
ENST00000367549.3 |
DHX9 |
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr21_-_26979786 | 0.85 |
ENST00000419219.1 ENST00000352957.4 ENST00000307301.7 |
MRPL39 |
mitochondrial ribosomal protein L39 |
| chrX_+_101854096 | 0.84 |
ENST00000246174.2 ENST00000537008.1 ENST00000541409.1 |
ARMCX5 |
armadillo repeat containing, X-linked 5 |
| chr12_+_53835539 | 0.84 |
ENST00000547368.1 ENST00000379786.4 ENST00000551945.1 |
PRR13 |
proline rich 13 |
| chr12_+_53835383 | 0.81 |
ENST00000429243.2 |
PRR13 |
proline rich 13 |
| chr7_-_44613494 | 0.78 |
ENST00000431640.1 ENST00000258772.5 |
DDX56 |
DEAD (Asp-Glu-Ala-Asp) box helicase 56 |
| chr4_-_2243839 | 0.77 |
ENST00000511885.2 ENST00000506763.1 ENST00000514395.1 ENST00000502440.1 ENST00000243706.4 ENST00000443786.2 |
POLN HAUS3 |
polymerase (DNA directed) nu HAUS augmin-like complex, subunit 3 |
| chr8_-_99129338 | 0.74 |
ENST00000520507.1 |
HRSP12 |
heat-responsive protein 12 |
| chr11_+_117857063 | 0.73 |
ENST00000227752.3 ENST00000541785.1 ENST00000545409.1 |
IL10RA |
interleukin 10 receptor, alpha |
| chr3_+_39424828 | 0.70 |
ENST00000273158.4 ENST00000431510.1 |
SLC25A38 |
solute carrier family 25, member 38 |
| chr8_-_99129384 | 0.68 |
ENST00000521560.1 ENST00000254878.3 |
HRSP12 |
heat-responsive protein 12 |
| chr7_+_99006232 | 0.65 |
ENST00000403633.2 |
BUD31 |
BUD31 homolog (S. cerevisiae) |
| chr9_-_123676827 | 0.59 |
ENST00000546084.1 |
TRAF1 |
TNF receptor-associated factor 1 |
| chr8_-_59572093 | 0.59 |
ENST00000427130.2 |
NSMAF |
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr10_+_94352956 | 0.57 |
ENST00000260731.3 |
KIF11 |
kinesin family member 11 |
| chrX_+_101854426 | 0.56 |
ENST00000536530.1 ENST00000473968.1 ENST00000604957.1 ENST00000477663.1 ENST00000479502.1 |
ARMCX5 |
armadillo repeat containing, X-linked 5 |
| chr1_+_26869597 | 0.55 |
ENST00000530003.1 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr17_+_80009741 | 0.52 |
ENST00000578552.1 ENST00000320548.4 ENST00000581578.1 ENST00000583885.1 ENST00000583641.1 ENST00000581418.1 ENST00000355130.2 ENST00000306823.6 ENST00000392358.2 |
GPS1 |
G protein pathway suppressor 1 |
| chr11_+_4116005 | 0.51 |
ENST00000300738.5 |
RRM1 |
ribonucleotide reductase M1 |
| chr14_+_35591858 | 0.49 |
ENST00000603544.1 |
KIAA0391 |
KIAA0391 |
| chr10_+_64893039 | 0.48 |
ENST00000277746.6 ENST00000435510.2 |
NRBF2 |
nuclear receptor binding factor 2 |
| chr11_+_4116054 | 0.48 |
ENST00000423050.2 |
RRM1 |
ribonucleotide reductase M1 |
| chr5_+_154238149 | 0.47 |
ENST00000519430.1 ENST00000520671.1 ENST00000521583.1 ENST00000518028.1 ENST00000519404.1 ENST00000519394.1 ENST00000518775.1 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr6_-_43655511 | 0.47 |
ENST00000372133.3 ENST00000372116.1 ENST00000427312.1 |
MRPS18A |
mitochondrial ribosomal protein S18A |
| chr18_-_11148587 | 0.45 |
ENST00000302079.6 ENST00000580640.1 ENST00000503781.3 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
| chr8_-_82024290 | 0.43 |
ENST00000220597.4 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr4_-_53525406 | 0.42 |
ENST00000451218.2 ENST00000441222.3 |
USP46 |
ubiquitin specific peptidase 46 |
| chr18_+_32558208 | 0.40 |
ENST00000436190.2 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr9_-_116102530 | 0.40 |
ENST00000374195.3 ENST00000341761.4 |
WDR31 |
WD repeat domain 31 |
| chr8_+_24151553 | 0.39 |
ENST00000265769.4 ENST00000540823.1 ENST00000397649.3 |
ADAM28 |
ADAM metallopeptidase domain 28 |
| chr12_+_53835508 | 0.38 |
ENST00000551003.1 ENST00000549068.1 ENST00000549740.1 ENST00000546581.1 ENST00000549581.1 ENST00000541275.1 |
PRR13 PCBP2 |
proline rich 13 poly(rC) binding protein 2 |
| chr2_+_95691445 | 0.37 |
ENST00000353004.3 ENST00000354078.3 ENST00000349807.3 |
MAL |
mal, T-cell differentiation protein |
| chr22_-_19166343 | 0.36 |
ENST00000215882.5 |
SLC25A1 |
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr3_+_196594727 | 0.35 |
ENST00000445299.2 ENST00000323460.5 ENST00000419026.1 |
SENP5 |
SUMO1/sentrin specific peptidase 5 |
| chr1_-_151319283 | 0.34 |
ENST00000392746.3 |
RFX5 |
regulatory factor X, 5 (influences HLA class II expression) |
| chr2_+_95691417 | 0.34 |
ENST00000309988.4 |
MAL |
mal, T-cell differentiation protein |
| chr8_+_24151620 | 0.34 |
ENST00000437154.2 |
ADAM28 |
ADAM metallopeptidase domain 28 |
| chr5_-_77590480 | 0.33 |
ENST00000519295.1 ENST00000255194.6 |
AP3B1 |
adaptor-related protein complex 3, beta 1 subunit |
| chr7_+_148936732 | 0.33 |
ENST00000335870.2 |
ZNF212 |
zinc finger protein 212 |
| chr15_-_55489097 | 0.32 |
ENST00000260443.4 |
RSL24D1 |
ribosomal L24 domain containing 1 |
| chrX_+_64887512 | 0.32 |
ENST00000360270.5 |
MSN |
moesin |
| chr15_-_25684110 | 0.31 |
ENST00000232165.3 |
UBE3A |
ubiquitin protein ligase E3A |
| chr2_+_27237615 | 0.30 |
ENST00000458529.1 ENST00000402218.1 |
MAPRE3 |
microtubule-associated protein, RP/EB family, member 3 |
| chr10_+_88428370 | 0.29 |
ENST00000372066.3 ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3 |
LIM domain binding 3 |
| chr2_+_89890533 | 0.29 |
ENST00000429992.2 |
IGKV2D-40 |
immunoglobulin kappa variable 2D-40 |
| chr5_+_154238096 | 0.29 |
ENST00000517568.1 ENST00000524105.1 ENST00000285896.6 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr19_-_59070239 | 0.29 |
ENST00000595957.1 ENST00000253023.3 |
UBE2M |
ubiquitin-conjugating enzyme E2M |
| chr5_+_154238042 | 0.28 |
ENST00000519211.1 ENST00000522458.1 ENST00000519903.1 ENST00000521450.1 ENST00000403027.2 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr14_+_35591928 | 0.23 |
ENST00000605870.1 ENST00000557404.3 |
KIAA0391 |
KIAA0391 |
| chr19_+_18669809 | 0.21 |
ENST00000602094.1 |
KXD1 |
KxDL motif containing 1 |
| chr10_+_88428206 | 0.19 |
ENST00000429277.2 ENST00000458213.2 ENST00000352360.5 |
LDB3 |
LIM domain binding 3 |
| chr22_-_29196030 | 0.19 |
ENST00000405219.3 |
XBP1 |
X-box binding protein 1 |
| chr19_-_6481776 | 0.18 |
ENST00000543576.1 ENST00000590173.1 ENST00000381480.2 |
DENND1C |
DENN/MADD domain containing 1C |
| chr8_+_99129513 | 0.18 |
ENST00000522319.1 ENST00000401707.2 |
POP1 |
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr5_+_154237778 | 0.17 |
ENST00000523698.1 ENST00000517876.1 ENST00000520472.1 |
CNOT8 |
CCR4-NOT transcription complex, subunit 8 |
| chr14_+_79746249 | 0.17 |
ENST00000428277.2 |
NRXN3 |
neurexin 3 |
| chr3_+_52232102 | 0.16 |
ENST00000469224.1 ENST00000394965.2 ENST00000310271.2 ENST00000484952.1 |
ALAS1 |
aminolevulinate, delta-, synthase 1 |
| chr19_+_17862274 | 0.11 |
ENST00000596536.1 ENST00000593870.1 ENST00000598086.1 ENST00000598932.1 ENST00000595023.1 ENST00000594068.1 ENST00000596507.1 ENST00000595033.1 ENST00000597718.1 |
FCHO1 |
FCH domain only 1 |
| chrX_-_153210107 | 0.09 |
ENST00000369997.3 ENST00000393700.3 ENST00000412763.1 |
RENBP |
renin binding protein |
| chr10_-_12084770 | 0.08 |
ENST00000357604.5 |
UPF2 |
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr14_-_35591433 | 0.08 |
ENST00000261475.5 ENST00000555644.1 |
PPP2R3C |
protein phosphatase 2, regulatory subunit B'', gamma |
| chr16_-_28621353 | 0.06 |
ENST00000567512.1 |
SULT1A1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr22_-_29196546 | 0.06 |
ENST00000403532.3 ENST00000216037.6 |
XBP1 |
X-box binding protein 1 |
| chr1_+_8021954 | 0.06 |
ENST00000377491.1 ENST00000377488.1 |
PARK7 |
parkinson protein 7 |
| chr9_-_97401782 | 0.06 |
ENST00000375326.4 |
FBP1 |
fructose-1,6-bisphosphatase 1 |
| chr14_+_35591735 | 0.04 |
ENST00000604948.1 ENST00000605201.1 ENST00000250377.7 ENST00000321130.10 ENST00000534898.4 |
KIAA0391 |
KIAA0391 |
| chr17_-_3195876 | 0.03 |
ENST00000323404.1 |
OR3A1 |
olfactory receptor, family 3, subfamily A, member 1 |
| chr2_-_48982708 | 0.02 |
ENST00000428232.1 ENST00000405626.1 ENST00000294954.7 |
LHCGR |
luteinizing hormone/choriogonadotropin receptor |
| chr15_+_93749295 | 0.02 |
ENST00000599897.1 |
AC112693.2 |
AC112693.2 |
| chr20_-_32700075 | 0.02 |
ENST00000374980.2 |
EIF2S2 |
eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.4 | 1.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.3 | 1.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 1.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 1.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.6 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.7 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 0.4 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.4 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 0.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.4 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.1 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.2 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.8 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.8 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.2 | 1.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 0.8 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.1 | 1.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.5 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 2.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 1.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.3 | 1.3 | GO:0097476 | NMDA glutamate receptor clustering(GO:0097114) spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.2 | 1.0 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.2 | 1.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.8 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.3 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.4 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.7 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.1 | 0.7 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 0.2 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) regulation of lactation(GO:1903487) |
| 0.1 | 0.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 0.6 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.7 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.3 | GO:0051138 | positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 0.3 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 0.6 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.4 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.3 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 1.2 | GO:0061014 | positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 0.6 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0006050 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.5 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.3 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.0 | 1.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.8 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.1 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.2 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 1.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |