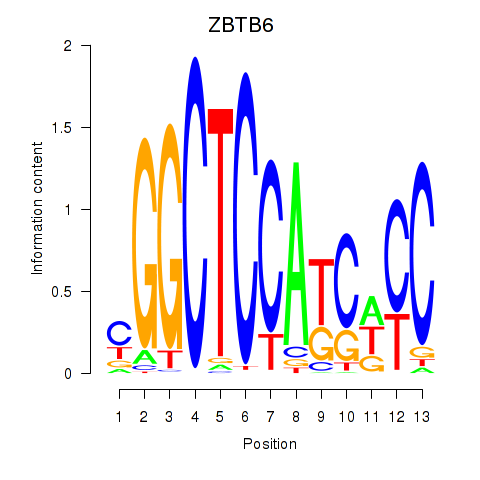
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for ZBTB6
Z-value: 1.35
Transcription factors associated with ZBTB6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB6
|
ENSG00000186130.4 | ZBTB6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB6 | hg19_v2_chr9_-_125675576_125675612 | 0.01 | 9.7e-01 | Click! |
Activity profile of ZBTB6 motif
Sorted Z-values of ZBTB6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_52887034 | 6.50 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chr12_-_52845910 | 5.12 |
ENST00000252252.3 |
KRT6B |
keratin 6B |
| chr10_+_47746929 | 2.88 |
ENST00000340243.6 ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2 AL603965.1 |
annexin A8-like 2 Protein LOC100996760 |
| chr10_+_48255253 | 2.84 |
ENST00000357718.4 ENST00000344416.5 ENST00000456111.2 ENST00000374258.3 |
ANXA8 AL591684.1 |
annexin A8 Protein LOC100996760 |
| chr10_-_47173994 | 2.71 |
ENST00000414655.2 ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1 LINC00842 |
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chrX_+_131157322 | 2.56 |
ENST00000481105.1 ENST00000354719.6 ENST00000394335.2 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chrX_+_131157290 | 2.56 |
ENST00000394334.2 |
MST4 |
Serine/threonine-protein kinase MST4 |
| chr1_+_26856236 | 2.56 |
ENST00000374168.2 ENST00000374166.4 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr2_-_20424844 | 2.54 |
ENST00000403076.1 ENST00000254351.4 |
SDC1 |
syndecan 1 |
| chr18_+_29077990 | 2.46 |
ENST00000261590.8 |
DSG2 |
desmoglein 2 |
| chr16_+_22825475 | 2.15 |
ENST00000261374.3 |
HS3ST2 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
| chr1_-_153538292 | 2.06 |
ENST00000497140.1 ENST00000368708.3 |
S100A2 |
S100 calcium binding protein A2 |
| chr1_-_153538011 | 2.06 |
ENST00000368707.4 |
S100A2 |
S100 calcium binding protein A2 |
| chr17_-_39677971 | 1.94 |
ENST00000393976.2 |
KRT15 |
keratin 15 |
| chr12_-_53012343 | 1.80 |
ENST00000305748.3 |
KRT73 |
keratin 73 |
| chr17_+_48609903 | 1.69 |
ENST00000268933.3 |
EPN3 |
epsin 3 |
| chr12_-_52967600 | 1.61 |
ENST00000549343.1 ENST00000305620.2 |
KRT74 |
keratin 74 |
| chr22_+_23229960 | 1.32 |
ENST00000526893.1 ENST00000532223.2 ENST00000531372.1 |
IGLL5 |
immunoglobulin lambda-like polypeptide 5 |
| chr8_+_104513086 | 1.25 |
ENST00000406091.3 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr1_+_152957707 | 1.23 |
ENST00000368762.1 |
SPRR1A |
small proline-rich protein 1A |
| chr3_-_189838670 | 1.14 |
ENST00000319332.5 |
LEPREL1 |
leprecan-like 1 |
| chr1_-_108507631 | 1.04 |
ENST00000527011.1 ENST00000370056.4 |
VAV3 |
vav 3 guanine nucleotide exchange factor |
| chr9_-_123691047 | 0.99 |
ENST00000373887.3 |
TRAF1 |
TNF receptor-associated factor 1 |
| chr15_+_43985725 | 0.96 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr6_+_29691056 | 0.96 |
ENST00000414333.1 ENST00000334668.4 ENST00000259951.7 |
HLA-F |
major histocompatibility complex, class I, F |
| chr10_-_121296045 | 0.90 |
ENST00000392865.1 |
RGS10 |
regulator of G-protein signaling 10 |
| chr15_+_43886057 | 0.87 |
ENST00000441322.1 ENST00000413657.2 ENST00000453733.1 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr6_+_30850697 | 0.87 |
ENST00000509639.1 ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr16_+_23847339 | 0.85 |
ENST00000303531.7 |
PRKCB |
protein kinase C, beta |
| chr1_-_93426998 | 0.84 |
ENST00000370310.4 |
FAM69A |
family with sequence similarity 69, member A |
| chr17_+_15848231 | 0.82 |
ENST00000304222.2 |
ADORA2B |
adenosine A2b receptor |
| chrX_+_73641286 | 0.78 |
ENST00000587091.1 |
SLC16A2 |
solute carrier family 16, member 2 (thyroid hormone transporter) |
| chrX_+_13707235 | 0.78 |
ENST00000464506.1 |
RAB9A |
RAB9A, member RAS oncogene family |
| chr22_-_46933067 | 0.77 |
ENST00000262738.3 ENST00000395964.1 |
CELSR1 |
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr6_+_149068464 | 0.77 |
ENST00000367463.4 |
UST |
uronyl-2-sulfotransferase |
| chr12_+_56325812 | 0.74 |
ENST00000394147.1 ENST00000551156.1 ENST00000553783.1 ENST00000557080.1 ENST00000432422.3 ENST00000556001.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr22_-_23922410 | 0.73 |
ENST00000249053.3 |
IGLL1 |
immunoglobulin lambda-like polypeptide 1 |
| chr10_-_105615164 | 0.72 |
ENST00000355946.2 ENST00000369774.4 |
SH3PXD2A |
SH3 and PX domains 2A |
| chr3_-_190040223 | 0.72 |
ENST00000295522.3 |
CLDN1 |
claudin 1 |
| chr9_-_123691439 | 0.66 |
ENST00000540010.1 |
TRAF1 |
TNF receptor-associated factor 1 |
| chr19_-_51529849 | 0.66 |
ENST00000600362.1 ENST00000453757.3 ENST00000601671.1 |
KLK11 |
kallikrein-related peptidase 11 |
| chr19_-_39108568 | 0.66 |
ENST00000586296.1 |
MAP4K1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr1_+_45965725 | 0.64 |
ENST00000401061.4 |
MMACHC |
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr17_+_48610074 | 0.64 |
ENST00000503690.1 ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3 |
epsin 3 |
| chr5_-_72744336 | 0.63 |
ENST00000499003.3 |
FOXD1 |
forkhead box D1 |
| chr22_+_43506747 | 0.63 |
ENST00000216115.2 |
BIK |
BCL2-interacting killer (apoptosis-inducing) |
| chr9_-_35650900 | 0.63 |
ENST00000259608.3 |
SIT1 |
signaling threshold regulating transmembrane adaptor 1 |
| chr4_+_1795012 | 0.62 |
ENST00000481110.2 ENST00000340107.4 ENST00000440486.2 ENST00000412135.2 |
FGFR3 |
fibroblast growth factor receptor 3 |
| chr18_+_42260861 | 0.61 |
ENST00000282030.5 |
SETBP1 |
SET binding protein 1 |
| chr16_+_50776021 | 0.61 |
ENST00000566679.2 ENST00000564634.1 ENST00000398568.2 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr1_-_204380919 | 0.61 |
ENST00000367188.4 |
PPP1R15B |
protein phosphatase 1, regulatory subunit 15B |
| chr15_+_45422178 | 0.60 |
ENST00000389037.3 ENST00000558322.1 |
DUOX1 |
dual oxidase 1 |
| chr8_+_99956662 | 0.59 |
ENST00000523368.1 ENST00000297565.4 ENST00000435298.2 |
OSR2 |
odd-skipped related transciption factor 2 |
| chr8_+_98656336 | 0.58 |
ENST00000336273.3 |
MTDH |
metadherin |
| chr19_-_14628645 | 0.58 |
ENST00000598235.1 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr5_+_17217669 | 0.56 |
ENST00000322611.3 |
BASP1 |
brain abundant, membrane attached signal protein 1 |
| chr15_+_71185148 | 0.56 |
ENST00000443425.2 ENST00000560755.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr19_-_14629224 | 0.54 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr6_-_30712313 | 0.54 |
ENST00000376377.2 ENST00000259874.5 |
IER3 |
immediate early response 3 |
| chrX_-_9001351 | 0.53 |
ENST00000362066.3 |
FAM9B |
family with sequence similarity 9, member B |
| chr6_+_29795595 | 0.52 |
ENST00000360323.6 ENST00000376818.3 ENST00000376815.3 |
HLA-G |
major histocompatibility complex, class I, G |
| chr5_+_67511524 | 0.52 |
ENST00000521381.1 ENST00000521657.1 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr5_+_50678921 | 0.52 |
ENST00000230658.7 |
ISL1 |
ISL LIM homeobox 1 |
| chr20_-_56285595 | 0.51 |
ENST00000395816.3 ENST00000347215.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr19_-_39108552 | 0.50 |
ENST00000591517.1 |
MAP4K1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr4_-_103682071 | 0.50 |
ENST00000505239.1 |
MANBA |
mannosidase, beta A, lysosomal |
| chrX_+_44732757 | 0.49 |
ENST00000377967.4 ENST00000536777.1 ENST00000382899.4 ENST00000543216.1 |
KDM6A |
lysine (K)-specific demethylase 6A |
| chr11_+_65686802 | 0.49 |
ENST00000376991.2 |
DRAP1 |
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr9_-_88969339 | 0.48 |
ENST00000375960.2 ENST00000375961.2 |
ZCCHC6 |
zinc finger, CCHC domain containing 6 |
| chr4_+_1795508 | 0.48 |
ENST00000260795.2 ENST00000352904.1 |
FGFR3 |
fibroblast growth factor receptor 3 |
| chr11_+_47236489 | 0.48 |
ENST00000256996.4 ENST00000378603.3 ENST00000378600.3 ENST00000378601.3 |
DDB2 |
damage-specific DNA binding protein 2, 48kDa |
| chr11_+_65687158 | 0.48 |
ENST00000532933.1 |
DRAP1 |
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr17_+_7184986 | 0.48 |
ENST00000317370.8 ENST00000571308.1 |
SLC2A4 |
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr9_-_88969303 | 0.47 |
ENST00000277141.6 ENST00000375963.3 |
ZCCHC6 |
zinc finger, CCHC domain containing 6 |
| chr16_+_23847267 | 0.47 |
ENST00000321728.7 |
PRKCB |
protein kinase C, beta |
| chr11_+_63753883 | 0.47 |
ENST00000538426.1 ENST00000543004.1 |
OTUB1 |
OTU domain, ubiquitin aldehyde binding 1 |
| chr2_-_25896380 | 0.47 |
ENST00000545439.1 ENST00000407186.1 ENST00000406818.3 ENST00000404103.3 ENST00000407661.3 ENST00000407038.3 ENST00000405222.1 ENST00000288642.8 |
DTNB |
dystrobrevin, beta |
| chr22_-_23922448 | 0.46 |
ENST00000438703.1 ENST00000330377.2 |
IGLL1 |
immunoglobulin lambda-like polypeptide 1 |
| chr12_-_6484376 | 0.46 |
ENST00000360168.3 ENST00000358945.3 |
SCNN1A |
sodium channel, non-voltage-gated 1 alpha subunit |
| chr16_+_50775971 | 0.45 |
ENST00000311559.9 ENST00000564326.1 ENST00000566206.1 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr3_+_112051994 | 0.45 |
ENST00000473539.1 ENST00000315711.8 ENST00000383681.3 |
CD200 |
CD200 molecule |
| chr2_+_62423242 | 0.45 |
ENST00000301998.4 |
B3GNT2 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 |
| chr16_+_15737124 | 0.45 |
ENST00000396355.1 ENST00000396353.2 |
NDE1 |
nudE neurodevelopment protein 1 |
| chr11_+_65686728 | 0.44 |
ENST00000312515.2 ENST00000525501.1 |
DRAP1 |
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr6_+_13574845 | 0.44 |
ENST00000606117.1 |
SIRT5 |
sirtuin 5 |
| chr8_+_98656693 | 0.44 |
ENST00000519934.1 |
MTDH |
metadherin |
| chr19_+_42817527 | 0.44 |
ENST00000598766.1 |
TMEM145 |
transmembrane protein 145 |
| chr22_+_45098067 | 0.43 |
ENST00000336985.6 ENST00000403696.1 ENST00000457960.1 ENST00000361473.5 |
PRR5 PRR5-ARHGAP8 |
proline rich 5 (renal) PRR5-ARHGAP8 readthrough |
| chrX_+_109246285 | 0.43 |
ENST00000372073.1 ENST00000372068.2 ENST00000288381.4 |
TMEM164 |
transmembrane protein 164 |
| chr11_+_65686952 | 0.43 |
ENST00000527119.1 |
DRAP1 |
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr16_+_30751953 | 0.42 |
ENST00000483578.1 |
RP11-2C24.4 |
RP11-2C24.4 |
| chr1_-_53793725 | 0.42 |
ENST00000371454.2 |
LRP8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr10_+_103825080 | 0.42 |
ENST00000299238.5 |
HPS6 |
Hermansky-Pudlak syndrome 6 |
| chr3_-_48470838 | 0.42 |
ENST00000358459.4 ENST00000358536.4 |
PLXNB1 |
plexin B1 |
| chr12_+_22199108 | 0.42 |
ENST00000229329.2 |
CMAS |
cytidine monophosphate N-acetylneuraminic acid synthetase |
| chr11_-_615570 | 0.42 |
ENST00000525445.1 ENST00000348655.6 ENST00000397566.1 |
IRF7 |
interferon regulatory factor 7 |
| chr9_-_72287191 | 0.41 |
ENST00000265381.4 |
APBA1 |
amyloid beta (A4) precursor protein-binding, family A, member 1 |
| chr8_+_56014949 | 0.41 |
ENST00000327381.6 |
XKR4 |
XK, Kell blood group complex subunit-related family, member 4 |
| chr5_-_140998616 | 0.41 |
ENST00000389054.3 ENST00000398562.2 ENST00000389057.5 ENST00000398566.3 ENST00000398557.4 ENST00000253811.6 |
DIAPH1 |
diaphanous-related formin 1 |
| chr15_+_81475047 | 0.41 |
ENST00000559388.1 |
IL16 |
interleukin 16 |
| chr15_+_78441663 | 0.41 |
ENST00000299518.2 ENST00000558554.1 ENST00000557826.1 ENST00000561279.1 ENST00000559186.1 ENST00000560770.1 ENST00000559881.1 ENST00000559205.1 ENST00000441490.2 |
IDH3A |
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr1_+_40627038 | 0.40 |
ENST00000372771.4 |
RLF |
rearranged L-myc fusion |
| chr14_+_50359773 | 0.39 |
ENST00000298316.5 |
ARF6 |
ADP-ribosylation factor 6 |
| chr17_-_29151794 | 0.39 |
ENST00000324238.6 |
CRLF3 |
cytokine receptor-like factor 3 |
| chr6_-_41040268 | 0.38 |
ENST00000373154.2 ENST00000244558.9 ENST00000464633.1 ENST00000424266.2 ENST00000479950.1 ENST00000482515.1 |
OARD1 |
O-acyl-ADP-ribose deacylase 1 |
| chr10_+_72575643 | 0.38 |
ENST00000373202.3 |
SGPL1 |
sphingosine-1-phosphate lyase 1 |
| chr1_-_53793584 | 0.38 |
ENST00000354412.3 ENST00000347547.2 ENST00000306052.6 |
LRP8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr11_-_119599794 | 0.38 |
ENST00000264025.3 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr12_+_48513009 | 0.37 |
ENST00000359794.5 ENST00000551339.1 ENST00000395233.2 ENST00000548345.1 |
PFKM |
phosphofructokinase, muscle |
| chr10_+_6244829 | 0.37 |
ENST00000317350.4 ENST00000379785.1 ENST00000379782.3 ENST00000360521.2 ENST00000379775.4 |
PFKFB3 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr6_+_41040678 | 0.37 |
ENST00000341376.6 ENST00000353205.5 |
NFYA |
nuclear transcription factor Y, alpha |
| chr9_+_136223414 | 0.36 |
ENST00000371964.4 |
SURF2 |
surfeit 2 |
| chr1_+_28199047 | 0.36 |
ENST00000373925.1 ENST00000328928.7 ENST00000373927.3 ENST00000427466.1 ENST00000442118.1 ENST00000373921.3 |
THEMIS2 |
thymocyte selection associated family member 2 |
| chr3_+_181429704 | 0.36 |
ENST00000431565.2 ENST00000325404.1 |
SOX2 |
SRY (sex determining region Y)-box 2 |
| chrX_+_109245863 | 0.36 |
ENST00000372072.3 |
TMEM164 |
transmembrane protein 164 |
| chr1_+_12185949 | 0.35 |
ENST00000413146.2 |
TNFRSF8 |
tumor necrosis factor receptor superfamily, member 8 |
| chr4_-_151936865 | 0.35 |
ENST00000535741.1 |
LRBA |
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr5_+_86564739 | 0.35 |
ENST00000456692.2 ENST00000512763.1 ENST00000506290.1 |
RASA1 |
RAS p21 protein activator (GTPase activating protein) 1 |
| chr20_+_61448376 | 0.34 |
ENST00000343916.3 |
COL9A3 |
collagen, type IX, alpha 3 |
| chr22_+_31489344 | 0.34 |
ENST00000404574.1 |
SMTN |
smoothelin |
| chr1_-_54304212 | 0.34 |
ENST00000540001.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr2_-_38604398 | 0.34 |
ENST00000443098.1 ENST00000449130.1 ENST00000378954.4 ENST00000539122.1 ENST00000419554.2 ENST00000451483.1 ENST00000406122.1 |
ATL2 |
atlastin GTPase 2 |
| chr19_-_39108643 | 0.34 |
ENST00000396857.2 |
MAP4K1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr16_-_31021717 | 0.34 |
ENST00000565419.1 |
STX1B |
syntaxin 1B |
| chr9_+_126773880 | 0.33 |
ENST00000373615.4 |
LHX2 |
LIM homeobox 2 |
| chr12_-_92539614 | 0.33 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr1_-_54303949 | 0.33 |
ENST00000234725.8 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr11_-_615942 | 0.32 |
ENST00000397562.3 ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7 |
interferon regulatory factor 7 |
| chr16_-_31021921 | 0.32 |
ENST00000215095.5 |
STX1B |
syntaxin 1B |
| chr1_-_236445251 | 0.32 |
ENST00000354619.5 ENST00000327333.8 |
ERO1LB |
ERO1-like beta (S. cerevisiae) |
| chr7_+_98972345 | 0.31 |
ENST00000418347.2 ENST00000429246.1 ENST00000417330.1 ENST00000431816.1 ENST00000427217.1 ENST00000458033.1 ENST00000451682.1 |
ARPC1B |
actin related protein 2/3 complex, subunit 1B, 41kDa |
| chr6_+_29910301 | 0.31 |
ENST00000376809.5 ENST00000376802.2 |
HLA-A |
major histocompatibility complex, class I, A |
| chr7_+_145813453 | 0.30 |
ENST00000361727.3 |
CNTNAP2 |
contactin associated protein-like 2 |
| chr16_+_29827832 | 0.30 |
ENST00000609618.1 |
PAGR1 |
PAXIP1-associated glutamate-rich protein 1 |
| chr16_+_50775948 | 0.30 |
ENST00000569681.1 ENST00000569418.1 ENST00000540145.1 |
CYLD |
cylindromatosis (turban tumor syndrome) |
| chr22_+_22764088 | 0.30 |
ENST00000390299.2 |
IGLV1-40 |
immunoglobulin lambda variable 1-40 |
| chr3_+_14989186 | 0.30 |
ENST00000435454.1 ENST00000323373.6 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr5_-_140998481 | 0.30 |
ENST00000518047.1 |
DIAPH1 |
diaphanous-related formin 1 |
| chr13_+_28712614 | 0.30 |
ENST00000380958.3 |
PAN3 |
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr7_+_98972298 | 0.30 |
ENST00000252725.5 |
ARPC1B |
actin related protein 2/3 complex, subunit 1B, 41kDa |
| chr5_+_131892603 | 0.30 |
ENST00000378823.3 ENST00000265335.6 |
RAD50 |
RAD50 homolog (S. cerevisiae) |
| chr3_+_38206975 | 0.29 |
ENST00000446845.1 ENST00000311806.3 |
OXSR1 |
oxidative stress responsive 1 |
| chr10_+_103912137 | 0.29 |
ENST00000603742.1 ENST00000488254.2 ENST00000461421.1 ENST00000476468.1 ENST00000370007.5 |
NOLC1 |
nucleolar and coiled-body phosphoprotein 1 |
| chr6_+_87865262 | 0.29 |
ENST00000369577.3 ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292 |
zinc finger protein 292 |
| chr14_+_72052983 | 0.29 |
ENST00000358550.2 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
| chr2_+_166095898 | 0.29 |
ENST00000424833.1 ENST00000375437.2 ENST00000357398.3 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr9_+_71320596 | 0.29 |
ENST00000265382.3 |
PIP5K1B |
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr3_-_27525865 | 0.28 |
ENST00000445684.1 |
SLC4A7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr7_-_98741642 | 0.28 |
ENST00000361368.2 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
| chr7_-_100823496 | 0.28 |
ENST00000455377.1 ENST00000443096.1 ENST00000300303.2 |
NAT16 |
N-acetyltransferase 16 (GCN5-related, putative) |
| chr12_-_133707021 | 0.28 |
ENST00000537226.1 |
ZNF891 |
zinc finger protein 891 |
| chr22_-_51017084 | 0.28 |
ENST00000360719.2 ENST00000457250.1 ENST00000440709.1 |
CPT1B |
carnitine palmitoyltransferase 1B (muscle) |
| chr3_-_27525826 | 0.27 |
ENST00000454389.1 ENST00000440156.1 ENST00000437179.1 ENST00000446700.1 ENST00000455077.1 ENST00000435667.2 ENST00000388777.4 ENST00000425128.2 |
SLC4A7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr10_-_27149904 | 0.27 |
ENST00000376166.1 ENST00000376138.3 ENST00000355394.4 ENST00000346832.5 ENST00000376134.3 ENST00000376137.4 ENST00000536334.1 ENST00000490841.2 |
ABI1 |
abl-interactor 1 |
| chr18_+_55816546 | 0.27 |
ENST00000435432.2 ENST00000357895.5 ENST00000586263.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr19_+_30302805 | 0.27 |
ENST00000262643.3 ENST00000575243.1 ENST00000357943.5 |
CCNE1 |
cyclin E1 |
| chr10_-_103880209 | 0.27 |
ENST00000425280.1 |
LDB1 |
LIM domain binding 1 |
| chr8_-_80942061 | 0.27 |
ENST00000519386.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr20_-_34287103 | 0.27 |
ENST00000374085.1 ENST00000419569.1 |
NFS1 |
NFS1 cysteine desulfurase |
| chr11_+_63606558 | 0.26 |
ENST00000350490.7 ENST00000502399.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr4_+_56815102 | 0.26 |
ENST00000257287.4 |
CEP135 |
centrosomal protein 135kDa |
| chr10_+_103911926 | 0.25 |
ENST00000605788.1 ENST00000405356.1 |
NOLC1 |
nucleolar and coiled-body phosphoprotein 1 |
| chr17_+_77893135 | 0.25 |
ENST00000574526.1 ENST00000572353.1 |
RP11-353N14.4 |
RP11-353N14.4 |
| chr9_+_128509624 | 0.25 |
ENST00000342287.5 ENST00000373487.4 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
| chr8_-_72756667 | 0.25 |
ENST00000325509.4 |
MSC |
musculin |
| chr11_+_71846764 | 0.25 |
ENST00000456237.1 ENST00000442948.2 ENST00000546166.1 |
FOLR3 |
folate receptor 3 (gamma) |
| chr15_+_64752927 | 0.25 |
ENST00000416172.1 |
ZNF609 |
zinc finger protein 609 |
| chr14_-_106967788 | 0.24 |
ENST00000390622.2 |
IGHV1-46 |
immunoglobulin heavy variable 1-46 |
| chr4_+_56814968 | 0.24 |
ENST00000422247.2 |
CEP135 |
centrosomal protein 135kDa |
| chr3_+_133292851 | 0.24 |
ENST00000503932.1 |
CDV3 |
CDV3 homolog (mouse) |
| chr8_-_99837856 | 0.24 |
ENST00000518165.1 ENST00000419617.2 |
STK3 |
serine/threonine kinase 3 |
| chr7_+_4721885 | 0.24 |
ENST00000328914.4 |
FOXK1 |
forkhead box K1 |
| chr2_-_172017393 | 0.24 |
ENST00000442919.2 |
TLK1 |
tousled-like kinase 1 |
| chr3_-_120068143 | 0.24 |
ENST00000295628.3 |
LRRC58 |
leucine rich repeat containing 58 |
| chr3_+_48507621 | 0.24 |
ENST00000456089.1 |
TREX1 |
three prime repair exonuclease 1 |
| chr10_-_27149851 | 0.24 |
ENST00000376142.2 ENST00000359188.4 ENST00000376139.2 ENST00000376160.1 |
ABI1 |
abl-interactor 1 |
| chr8_-_117768023 | 0.24 |
ENST00000518949.1 ENST00000522453.1 ENST00000521861.1 ENST00000518995.1 |
EIF3H |
eukaryotic translation initiation factor 3, subunit H |
| chr10_-_27149792 | 0.23 |
ENST00000376140.3 ENST00000376170.4 |
ABI1 |
abl-interactor 1 |
| chr14_-_50583271 | 0.23 |
ENST00000395860.2 ENST00000395859.2 |
VCPKMT |
valosin containing protein lysine (K) methyltransferase |
| chr16_+_66968343 | 0.23 |
ENST00000417689.1 ENST00000561697.1 ENST00000317091.4 ENST00000566182.1 |
CES2 |
carboxylesterase 2 |
| chr8_-_80942467 | 0.23 |
ENST00000518271.1 ENST00000276585.4 ENST00000521605.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr19_+_47104553 | 0.23 |
ENST00000598871.1 ENST00000594523.1 |
CALM3 |
calmodulin 3 (phosphorylase kinase, delta) |
| chr8_-_80942139 | 0.23 |
ENST00000521434.1 ENST00000519120.1 ENST00000520946.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr16_+_56716336 | 0.23 |
ENST00000394485.4 ENST00000562939.1 |
MT1X |
metallothionein 1X |
| chr11_+_119076745 | 0.23 |
ENST00000264033.4 |
CBL |
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr4_+_30723003 | 0.22 |
ENST00000543491.1 |
PCDH7 |
protocadherin 7 |
| chr8_-_21646311 | 0.22 |
ENST00000524240.1 ENST00000400782.4 |
GFRA2 |
GDNF family receptor alpha 2 |
| chr3_+_128720424 | 0.22 |
ENST00000480450.1 ENST00000436022.2 |
EFCC1 |
EF-hand and coiled-coil domain containing 1 |
| chr2_-_37193606 | 0.22 |
ENST00000379213.2 ENST00000263918.4 |
STRN |
striatin, calmodulin binding protein |
| chr22_-_51016846 | 0.22 |
ENST00000312108.7 ENST00000395650.2 |
CPT1B |
carnitine palmitoyltransferase 1B (muscle) |
| chr1_-_92949505 | 0.22 |
ENST00000370332.1 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr1_+_26737253 | 0.22 |
ENST00000326279.6 |
LIN28A |
lin-28 homolog A (C. elegans) |
| chr17_+_7608511 | 0.22 |
ENST00000226091.2 |
EFNB3 |
ephrin-B3 |
| chr15_-_80215984 | 0.22 |
ENST00000485386.1 ENST00000479961.1 |
ST20 ST20-MTHFS |
suppressor of tumorigenicity 20 ST20-MTHFS readthrough |
| chr14_-_64010046 | 0.22 |
ENST00000337537.3 |
PPP2R5E |
protein phosphatase 2, regulatory subunit B', epsilon isoform |
| chr7_-_98741714 | 0.22 |
ENST00000361125.1 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
| chr8_+_67341239 | 0.22 |
ENST00000320270.2 |
RRS1 |
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr20_-_44485835 | 0.21 |
ENST00000457981.1 ENST00000426915.1 ENST00000217455.4 |
ACOT8 |
acyl-CoA thioesterase 8 |
| chr4_-_102268484 | 0.21 |
ENST00000394853.4 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chrX_-_153200676 | 0.21 |
ENST00000464845.1 |
NAA10 |
N(alpha)-acetyltransferase 10, NatA catalytic subunit |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 3.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 5.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 4.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 5.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 1.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.4 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.9 | 2.8 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.8 | 2.5 | GO:0048627 | myoblast development(GO:0048627) |
| 0.5 | 2.5 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.5 | 1.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.4 | 1.3 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.4 | 2.6 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.3 | 0.6 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.3 | 0.8 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.2 | 0.2 | GO:2000291 | myoblast proliferation(GO:0051450) regulation of myoblast proliferation(GO:2000291) |
| 0.2 | 0.7 | GO:1903348 | cellular response to lead ion(GO:0071284) positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.2 | 0.7 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.2 | 0.7 | GO:0070631 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.2 | 1.9 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.2 | 0.8 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.2 | 0.7 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.2 | 1.8 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.2 | 0.4 | GO:0032759 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.2 | 0.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.2 | 0.5 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.2 | 0.8 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 0.2 | GO:1903525 | regulation of membrane tubulation(GO:1903525) positive regulation of membrane tubulation(GO:1903527) |
| 0.2 | 4.9 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.2 | 0.5 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 0.6 | GO:0042335 | cuticle development(GO:0042335) |
| 0.1 | 0.7 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.1 | 0.1 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 | 0.4 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.4 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 | 0.6 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.1 | 0.4 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.1 | 0.6 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.1 | 0.3 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.1 | 0.6 | GO:2001076 | regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.1 | 0.8 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.1 | 0.4 | GO:0018002 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.1 | 0.3 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.3 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.1 | 0.3 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.1 | 0.3 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.3 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.7 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.9 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 1.5 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.8 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.1 | 0.6 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 0.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.3 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 0.2 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.1 | 0.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.1 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.1 | 0.2 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 0.2 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.2 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.1 | 0.6 | GO:1903912 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.1 | 0.2 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.2 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.1 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 7.1 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.2 | GO:1900060 | negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 0.8 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.0 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) L-kynurenine metabolic process(GO:0097052) |
| 0.1 | 0.2 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.1 | 0.3 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 0.5 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.3 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.2 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.1 | 2.8 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.1 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 0.9 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.4 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.0 | 0.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.8 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.5 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.5 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.6 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.2 | GO:0036018 | cellular response to erythropoietin(GO:0036018) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.2 | GO:0090649 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.4 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 2.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.3 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.3 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.4 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.2 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 0.0 | 0.1 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.2 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.4 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.1 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 0.2 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.4 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.1 | GO:0034971 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) histone H3-R17 methylation(GO:0034971) |
| 0.0 | 0.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.3 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:2000973 | midbrain morphogenesis(GO:1904693) regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.4 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.3 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.0 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.0 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 1.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.3 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.0 | GO:0015672 | monovalent inorganic cation transport(GO:0015672) |
| 0.0 | 0.2 | GO:0002870 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.5 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.2 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 1.2 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 1.0 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.0 | 1.8 | GO:0006023 | aminoglycan biosynthetic process(GO:0006023) glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.5 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 2.7 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 0.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 1.0 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.2 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.0 | GO:0060897 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.1 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 1.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.3 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.4 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.5 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.3 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 2.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 4.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 4.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 2.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.1 | 1.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.7 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.4 | 2.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 2.5 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.3 | 1.6 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 1.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.2 | 0.7 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 0.8 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.2 | 0.9 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 1.0 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.2 | 0.5 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 1.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 2.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.4 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.1 | 0.7 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.4 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 0.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 4.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.5 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.4 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 1.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 13.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.2 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 1.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.2 | GO:0031177 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.2 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.4 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 2.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.2 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 0.3 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 0.1 | 0.2 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.1 | 0.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 0.3 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.4 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.3 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0047374 | methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 2.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.6 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.3 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.4 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0004782 | sulfinoalanine decarboxylase activity(GO:0004782) |
| 0.0 | 0.6 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.8 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.6 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.0 | 0.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.7 | GO:0034979 | NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.0 | 0.2 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.8 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.2 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.0 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.0 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 1.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 15.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 0.7 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.2 | 1.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.5 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.3 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.4 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 2.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.3 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 2.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.1 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.5 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.4 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 2.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 2.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.4 | GO:0031082 | BLOC complex(GO:0031082) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 1.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 0.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |