Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
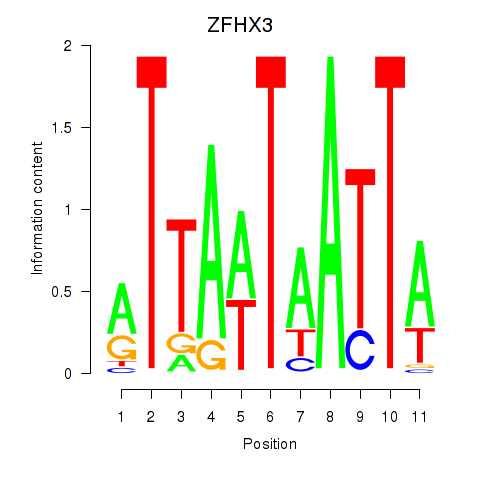
Results for ZFHX3
Z-value: 0.54
Transcription factors associated with ZFHX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZFHX3
|
ENSG00000140836.10 | ZFHX3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZFHX3 | hg19_v2_chr16_-_73093597_73093597, hg19_v2_chr16_-_73082274_73082274 | 0.29 | 2.7e-01 | Click! |
Activity profile of ZFHX3 motif
Sorted Z-values of ZFHX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZFHX3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_22696314 | 1.34 |
ENST00000532398.1 ENST00000433790.1 |
GAS2 |
growth arrest-specific 2 |
| chr4_-_155511887 | 0.92 |
ENST00000302053.3 ENST00000403106.3 |
FGA |
fibrinogen alpha chain |
| chr9_-_123812542 | 0.80 |
ENST00000223642.1 |
C5 |
complement component 5 |
| chr17_-_64225508 | 0.71 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr12_-_91546926 | 0.61 |
ENST00000550758.1 |
DCN |
decorin |
| chr5_+_118690466 | 0.58 |
ENST00000503646.1 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
| chr19_+_926000 | 0.57 |
ENST00000263620.3 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
| chrX_-_110655306 | 0.56 |
ENST00000371993.2 |
DCX |
doublecortin |
| chr17_+_41052808 | 0.55 |
ENST00000592383.1 ENST00000253801.2 ENST00000585489.1 |
G6PC |
glucose-6-phosphatase, catalytic subunit |
| chr1_+_192544857 | 0.52 |
ENST00000367459.3 ENST00000469578.2 |
RGS1 |
regulator of G-protein signaling 1 |
| chr14_+_95027772 | 0.47 |
ENST00000555095.1 ENST00000298841.5 ENST00000554220.1 ENST00000553780.1 |
SERPINA4 SERPINA5 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr4_+_100495864 | 0.44 |
ENST00000265517.5 ENST00000422897.2 |
MTTP |
microsomal triglyceride transfer protein |
| chr12_+_59989791 | 0.43 |
ENST00000552432.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr5_+_95066823 | 0.42 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr8_-_70745575 | 0.41 |
ENST00000524945.1 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr22_-_30970560 | 0.39 |
ENST00000402369.1 ENST00000406361.1 |
GAL3ST1 |
galactose-3-O-sulfotransferase 1 |
| chr10_-_75226166 | 0.36 |
ENST00000544628.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chrX_-_32173579 | 0.35 |
ENST00000359836.1 ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD |
dystrophin |
| chr19_-_46234119 | 0.32 |
ENST00000317683.3 |
FBXO46 |
F-box protein 46 |
| chr17_-_26697304 | 0.32 |
ENST00000536498.1 |
VTN |
vitronectin |
| chrX_+_9431324 | 0.32 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr2_-_60780607 | 0.32 |
ENST00000537768.1 ENST00000335712.6 ENST00000356842.4 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr2_-_60780702 | 0.32 |
ENST00000359629.5 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr19_-_36304201 | 0.31 |
ENST00000301175.3 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chr2_-_60780536 | 0.30 |
ENST00000538214.1 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr15_+_58724184 | 0.25 |
ENST00000433326.2 |
LIPC |
lipase, hepatic |
| chrX_+_84258832 | 0.21 |
ENST00000373173.2 |
APOOL |
apolipoprotein O-like |
| chr17_-_60142609 | 0.21 |
ENST00000397786.2 |
MED13 |
mediator complex subunit 13 |
| chr3_-_114477787 | 0.20 |
ENST00000464560.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr1_-_160549235 | 0.20 |
ENST00000368054.3 ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84 |
CD84 molecule |
| chr7_-_100026280 | 0.19 |
ENST00000360951.4 ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1 |
zinc finger, CW type with PWWP domain 1 |
| chr20_-_33735070 | 0.18 |
ENST00000374491.3 ENST00000542871.1 ENST00000374492.3 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr12_+_59989918 | 0.18 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr14_+_39703084 | 0.16 |
ENST00000553728.1 |
RP11-407N17.3 |
cTAGE family member 5 isoform 4 |
| chr8_+_98900132 | 0.16 |
ENST00000520016.1 |
MATN2 |
matrilin 2 |
| chr12_-_101801505 | 0.15 |
ENST00000539055.1 ENST00000551688.1 ENST00000551671.1 ENST00000261636.8 |
ARL1 |
ADP-ribosylation factor-like 1 |
| chr19_+_50016610 | 0.12 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr3_-_49066811 | 0.12 |
ENST00000442157.1 ENST00000326739.4 |
IMPDH2 |
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr6_-_116447283 | 0.11 |
ENST00000452729.1 ENST00000243222.4 |
COL10A1 |
collagen, type X, alpha 1 |
| chr6_+_28317685 | 0.10 |
ENST00000252211.2 ENST00000341464.5 ENST00000377255.3 |
ZKSCAN3 |
zinc finger with KRAB and SCAN domains 3 |
| chr19_+_54466179 | 0.10 |
ENST00000270458.2 |
CACNG8 |
calcium channel, voltage-dependent, gamma subunit 8 |
| chr7_-_44580861 | 0.09 |
ENST00000546276.1 ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1 |
NPC1-like 1 |
| chrX_+_1710484 | 0.09 |
ENST00000313871.3 ENST00000381261.3 |
AKAP17A |
A kinase (PRKA) anchor protein 17A |
| chr4_-_100356291 | 0.09 |
ENST00000476959.1 ENST00000482593.1 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr16_+_87636474 | 0.08 |
ENST00000284262.2 |
JPH3 |
junctophilin 3 |
| chr7_+_100026406 | 0.08 |
ENST00000414441.1 |
MEPCE |
methylphosphate capping enzyme |
| chr2_+_185463093 | 0.08 |
ENST00000302277.6 |
ZNF804A |
zinc finger protein 804A |
| chr20_-_21494654 | 0.07 |
ENST00000377142.4 |
NKX2-2 |
NK2 homeobox 2 |
| chr3_+_69812701 | 0.07 |
ENST00000472437.1 |
MITF |
microphthalmia-associated transcription factor |
| chr11_-_61687739 | 0.06 |
ENST00000531922.1 ENST00000301773.5 |
RAB3IL1 |
RAB3A interacting protein (rabin3)-like 1 |
| chr21_+_40824003 | 0.05 |
ENST00000452550.1 |
SH3BGR |
SH3 domain binding glutamic acid-rich protein |
| chr12_-_7818474 | 0.05 |
ENST00000229304.4 |
APOBEC1 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr12_+_53835508 | 0.05 |
ENST00000551003.1 ENST00000549068.1 ENST00000549740.1 ENST00000546581.1 ENST00000549581.1 ENST00000541275.1 |
PRR13 PCBP2 |
proline rich 13 poly(rC) binding protein 2 |
| chr22_+_39101728 | 0.04 |
ENST00000216044.5 ENST00000484657.1 |
GTPBP1 |
GTP binding protein 1 |
| chr16_-_15474904 | 0.03 |
ENST00000534094.1 |
NPIPA5 |
nuclear pore complex interacting protein family, member A5 |
| chr1_+_159557607 | 0.03 |
ENST00000255040.2 |
APCS |
amyloid P component, serum |
| chr12_-_2944184 | 0.01 |
ENST00000337508.4 |
NRIP2 |
nuclear receptor interacting protein 2 |
| chr11_+_8040739 | 0.01 |
ENST00000534099.1 |
TUB |
tubby bipartite transcription factor |
| chr4_-_100356551 | 0.00 |
ENST00000209665.4 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr20_+_42187608 | 0.00 |
ENST00000373100.1 |
SGK2 |
serum/glucocorticoid regulated kinase 2 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.6 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.3 | 0.8 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 1.2 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 0.5 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 0.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.5 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 0.6 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.6 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.2 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.4 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0021779 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.7 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.6 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.4 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.8 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0036024 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.3 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.6 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.7 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |