Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for ZIC1
Z-value: 0.51
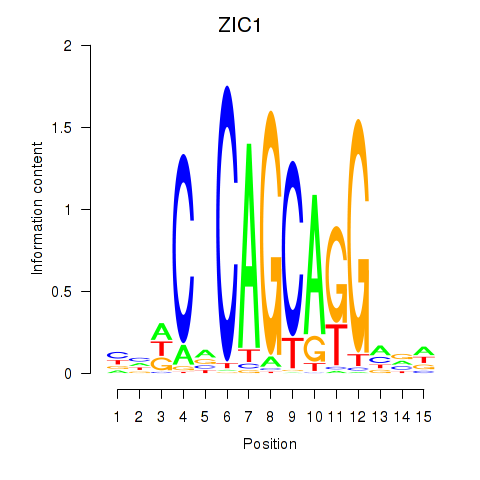
Transcription factors associated with ZIC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC1
|
ENSG00000152977.5 | ZIC1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZIC1 | hg19_v2_chr3_+_147127142_147127171 | -0.26 | 3.3e-01 | Click! |
Activity profile of ZIC1 motif
Sorted Z-values of ZIC1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_78622805 | 1.09 |
ENST00000373298.2 |
ITM2A |
integral membrane protein 2A |
| chr19_-_3028354 | 0.89 |
ENST00000586422.1 |
TLE2 |
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr16_+_771663 | 0.74 |
ENST00000568916.1 |
FAM173A |
family with sequence similarity 173, member A |
| chr1_+_65775204 | 0.71 |
ENST00000371069.4 |
DNAJC6 |
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr1_+_233749739 | 0.65 |
ENST00000366621.3 |
KCNK1 |
potassium channel, subfamily K, member 1 |
| chr13_-_29069232 | 0.58 |
ENST00000282397.4 ENST00000541932.1 ENST00000539099.1 |
FLT1 |
fms-related tyrosine kinase 1 |
| chr4_-_109090106 | 0.55 |
ENST00000379951.2 |
LEF1 |
lymphoid enhancer-binding factor 1 |
| chr1_-_71513471 | 0.53 |
ENST00000370931.3 ENST00000356595.4 ENST00000306666.5 ENST00000370932.2 ENST00000351052.5 ENST00000414819.1 ENST00000370924.4 |
PTGER3 |
prostaglandin E receptor 3 (subtype EP3) |
| chr1_-_1711508 | 0.50 |
ENST00000378625.1 |
NADK |
NAD kinase |
| chr18_-_52989525 | 0.48 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr10_-_36813162 | 0.48 |
ENST00000440465.1 |
NAMPTL |
nicotinamide phosphoribosyltransferase-like |
| chr1_+_89990431 | 0.48 |
ENST00000330947.2 ENST00000358200.4 |
LRRC8B |
leucine rich repeat containing 8 family, member B |
| chr18_-_52989217 | 0.46 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr1_+_24120143 | 0.44 |
ENST00000374501.1 |
LYPLA2 |
lysophospholipase II |
| chr16_+_2521500 | 0.43 |
ENST00000293973.1 |
NTN3 |
netrin 3 |
| chr10_-_62493223 | 0.43 |
ENST00000373827.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chrX_+_134975753 | 0.43 |
ENST00000535938.1 |
SAGE1 |
sarcoma antigen 1 |
| chrX_+_134975858 | 0.42 |
ENST00000537770.1 |
SAGE1 |
sarcoma antigen 1 |
| chr17_-_40428359 | 0.42 |
ENST00000293328.3 |
STAT5B |
signal transducer and activator of transcription 5B |
| chr12_-_15038779 | 0.39 |
ENST00000228938.5 ENST00000539261.1 |
MGP |
matrix Gla protein |
| chr4_-_109089573 | 0.37 |
ENST00000265165.1 |
LEF1 |
lymphoid enhancer-binding factor 1 |
| chr6_+_44187242 | 0.36 |
ENST00000393844.1 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr1_+_100315613 | 0.36 |
ENST00000361915.3 |
AGL |
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr11_-_134123142 | 0.36 |
ENST00000392595.2 ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1 |
thymocyte nuclear protein 1 |
| chr16_+_2255841 | 0.34 |
ENST00000301725.7 |
MLST8 |
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr3_+_14444063 | 0.32 |
ENST00000454876.2 ENST00000360861.3 ENST00000416216.2 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chrX_+_46771711 | 0.32 |
ENST00000424392.1 ENST00000397189.1 |
PHF16 |
jade family PHD finger 3 |
| chr7_+_150264365 | 0.32 |
ENST00000255945.2 ENST00000461940.1 |
GIMAP4 |
GTPase, IMAP family member 4 |
| chr1_+_95699704 | 0.30 |
ENST00000370202.4 |
RWDD3 |
RWD domain containing 3 |
| chr1_+_100316041 | 0.30 |
ENST00000370165.3 ENST00000370163.3 ENST00000294724.4 |
AGL |
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr1_-_244615425 | 0.30 |
ENST00000366535.3 |
ADSS |
adenylosuccinate synthase |
| chr6_-_35656685 | 0.29 |
ENST00000539068.1 ENST00000540787.1 |
FKBP5 |
FK506 binding protein 5 |
| chrX_+_46771848 | 0.28 |
ENST00000218343.4 |
PHF16 |
jade family PHD finger 3 |
| chr7_+_107384579 | 0.27 |
ENST00000222597.2 ENST00000415884.2 |
CBLL1 |
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr3_-_58563094 | 0.27 |
ENST00000464064.1 |
FAM107A |
family with sequence similarity 107, member A |
| chr21_-_39870339 | 0.27 |
ENST00000429727.2 ENST00000398905.1 ENST00000398907.1 ENST00000453032.2 ENST00000288319.7 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr1_+_95699740 | 0.26 |
ENST00000429514.2 ENST00000263893.6 |
RWDD3 |
RWD domain containing 3 |
| chr1_+_46668994 | 0.26 |
ENST00000371980.3 |
LURAP1 |
leucine rich adaptor protein 1 |
| chr9_-_35103105 | 0.26 |
ENST00000452248.2 ENST00000356493.5 |
STOML2 |
stomatin (EPB72)-like 2 |
| chr9_+_132934835 | 0.26 |
ENST00000372398.3 |
NCS1 |
neuronal calcium sensor 1 |
| chr1_-_111991850 | 0.25 |
ENST00000411751.2 |
WDR77 |
WD repeat domain 77 |
| chr10_-_49701686 | 0.25 |
ENST00000417247.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr6_-_35656712 | 0.25 |
ENST00000357266.4 ENST00000542713.1 |
FKBP5 |
FK506 binding protein 5 |
| chr1_-_113249948 | 0.25 |
ENST00000339083.7 ENST00000369642.3 |
RHOC |
ras homolog family member C |
| chr1_+_93544791 | 0.24 |
ENST00000545708.1 ENST00000540243.1 ENST00000370298.4 |
MTF2 |
metal response element binding transcription factor 2 |
| chr1_+_93544821 | 0.23 |
ENST00000370303.4 |
MTF2 |
metal response element binding transcription factor 2 |
| chr14_+_57735614 | 0.22 |
ENST00000261558.3 |
AP5M1 |
adaptor-related protein complex 5, mu 1 subunit |
| chr2_+_234668894 | 0.22 |
ENST00000305208.5 ENST00000608383.1 ENST00000360418.3 |
UGT1A8 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr7_-_107880508 | 0.22 |
ENST00000425651.2 |
NRCAM |
neuronal cell adhesion molecule |
| chr21_+_45209394 | 0.22 |
ENST00000497547.1 |
RRP1 |
ribosomal RNA processing 1 |
| chr10_+_21823079 | 0.21 |
ENST00000377100.3 ENST00000377072.3 ENST00000446906.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr1_-_43833628 | 0.21 |
ENST00000413844.2 ENST00000372458.3 |
ELOVL1 |
ELOVL fatty acid elongase 1 |
| chr11_+_75526212 | 0.20 |
ENST00000356136.3 |
UVRAG |
UV radiation resistance associated |
| chr11_+_64002292 | 0.20 |
ENST00000426086.2 |
VEGFB |
vascular endothelial growth factor B |
| chr7_-_37026108 | 0.19 |
ENST00000396045.3 |
ELMO1 |
engulfment and cell motility 1 |
| chr19_+_41509851 | 0.18 |
ENST00000593831.1 ENST00000330446.5 |
CYP2B6 |
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr1_-_161087802 | 0.18 |
ENST00000368010.3 |
PFDN2 |
prefoldin subunit 2 |
| chr1_+_111992064 | 0.18 |
ENST00000483994.1 |
ATP5F1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 |
| chr11_+_64001962 | 0.18 |
ENST00000309422.2 |
VEGFB |
vascular endothelial growth factor B |
| chr22_+_22599075 | 0.17 |
ENST00000403807.3 |
VPREB1 |
pre-B lymphocyte 1 |
| chr7_-_128045984 | 0.17 |
ENST00000470772.1 ENST00000480861.1 ENST00000496200.1 |
IMPDH1 |
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
| chrX_-_129299638 | 0.17 |
ENST00000535724.1 ENST00000346424.2 |
AIFM1 |
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr2_+_46769798 | 0.17 |
ENST00000238738.4 |
RHOQ |
ras homolog family member Q |
| chr6_+_41755389 | 0.16 |
ENST00000398884.3 ENST00000398881.3 |
TOMM6 |
translocase of outer mitochondrial membrane 6 homolog (yeast) |
| chr1_-_113249678 | 0.16 |
ENST00000369633.2 ENST00000425265.2 ENST00000369632.2 ENST00000436685.2 |
RHOC |
ras homolog family member C |
| chr8_-_22526597 | 0.16 |
ENST00000519513.1 ENST00000276416.6 ENST00000520292.1 ENST00000522268.1 |
BIN3 |
bridging integrator 3 |
| chr17_-_39222131 | 0.15 |
ENST00000394015.2 |
KRTAP2-4 |
keratin associated protein 2-4 |
| chr3_+_49057876 | 0.15 |
ENST00000326912.4 |
NDUFAF3 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr10_+_21823243 | 0.15 |
ENST00000307729.7 ENST00000377091.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chrX_-_129299847 | 0.15 |
ENST00000319908.3 ENST00000287295.3 |
AIFM1 |
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chrX_+_118425471 | 0.15 |
ENST00000428222.1 |
RP5-1139I1.1 |
RP5-1139I1.1 |
| chr2_+_169926047 | 0.14 |
ENST00000428522.1 ENST00000450153.1 ENST00000421653.1 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
| chr19_-_11450249 | 0.14 |
ENST00000222120.3 |
RAB3D |
RAB3D, member RAS oncogene family |
| chr1_+_16370271 | 0.13 |
ENST00000375679.4 |
CLCNKB |
chloride channel, voltage-sensitive Kb |
| chr22_+_29702572 | 0.13 |
ENST00000407647.2 ENST00000416823.1 ENST00000428622.1 |
GAS2L1 |
growth arrest-specific 2 like 1 |
| chr1_-_113249734 | 0.13 |
ENST00000484054.3 ENST00000369636.2 ENST00000369637.1 ENST00000285735.2 ENST00000369638.2 |
RHOC |
ras homolog family member C |
| chr6_+_31540056 | 0.12 |
ENST00000418386.2 |
LTA |
lymphotoxin alpha |
| chr7_-_135412925 | 0.12 |
ENST00000354042.4 |
SLC13A4 |
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chrX_+_13707235 | 0.12 |
ENST00000464506.1 |
RAB9A |
RAB9A, member RAS oncogene family |
| chrX_+_77166172 | 0.11 |
ENST00000343533.5 ENST00000350425.4 ENST00000341514.6 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
| chr15_+_85523671 | 0.11 |
ENST00000310298.4 ENST00000557957.1 |
PDE8A |
phosphodiesterase 8A |
| chr1_-_20446020 | 0.11 |
ENST00000375105.3 |
PLA2G2D |
phospholipase A2, group IID |
| chrX_+_24711997 | 0.11 |
ENST00000379068.3 ENST00000379059.3 |
POLA1 |
polymerase (DNA directed), alpha 1, catalytic subunit |
| chr14_+_38677123 | 0.10 |
ENST00000267377.2 |
SSTR1 |
somatostatin receptor 1 |
| chr11_-_2193025 | 0.10 |
ENST00000333684.5 ENST00000381178.1 ENST00000381175.1 ENST00000352909.3 |
TH |
tyrosine hydroxylase |
| chr10_-_81708854 | 0.10 |
ENST00000372292.3 |
SFTPD |
surfactant protein D |
| chr1_-_111991908 | 0.10 |
ENST00000235090.5 |
WDR77 |
WD repeat domain 77 |
| chr2_-_27938593 | 0.10 |
ENST00000379677.2 |
AC074091.13 |
Uncharacterized protein |
| chr19_-_50169064 | 0.09 |
ENST00000593337.1 ENST00000598808.1 ENST00000600453.1 ENST00000593818.1 ENST00000597198.1 ENST00000601809.1 ENST00000377139.3 |
IRF3 |
interferon regulatory factor 3 |
| chr1_+_161087873 | 0.09 |
ENST00000368009.2 ENST00000368007.4 ENST00000368008.1 ENST00000392190.5 |
NIT1 |
nitrilase 1 |
| chr17_+_40913210 | 0.09 |
ENST00000253796.5 |
RAMP2 |
receptor (G protein-coupled) activity modifying protein 2 |
| chr19_+_34287751 | 0.08 |
ENST00000590771.1 ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
| chr5_-_64920115 | 0.08 |
ENST00000381018.3 ENST00000274327.7 |
TRIM23 |
tripartite motif containing 23 |
| chr12_-_27091183 | 0.08 |
ENST00000544548.1 ENST00000261191.7 ENST00000537336.1 |
ASUN |
asunder spermatogenesis regulator |
| chr17_+_28804380 | 0.08 |
ENST00000225724.5 ENST00000451249.2 ENST00000467337.2 ENST00000581721.1 ENST00000414833.2 |
GOSR1 |
golgi SNAP receptor complex member 1 |
| chrX_+_70364667 | 0.08 |
ENST00000536169.1 ENST00000395855.2 ENST00000374051.3 ENST00000358741.3 |
NLGN3 |
neuroligin 3 |
| chr15_+_77712993 | 0.07 |
ENST00000336216.4 ENST00000381714.3 ENST00000558651.1 |
HMG20A |
high mobility group 20A |
| chr19_-_50168962 | 0.07 |
ENST00000599223.1 ENST00000593922.1 ENST00000600022.1 ENST00000596765.1 ENST00000599144.1 ENST00000596822.1 ENST00000598108.1 ENST00000601373.1 ENST00000595034.1 ENST00000601291.1 |
IRF3 |
interferon regulatory factor 3 |
| chr22_-_24622080 | 0.07 |
ENST00000425408.1 |
GGT5 |
gamma-glutamyltransferase 5 |
| chr1_+_16348366 | 0.07 |
ENST00000375692.1 ENST00000420078.1 |
CLCNKA |
chloride channel, voltage-sensitive Ka |
| chr14_-_50999190 | 0.07 |
ENST00000557390.1 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr4_-_66536057 | 0.07 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chr5_+_64920543 | 0.06 |
ENST00000399438.3 ENST00000510585.2 |
TRAPPC13 CTC-534A2.2 |
trafficking protein particle complex 13 CDNA FLJ26957 fis, clone SLV00486; Uncharacterized protein |
| chr9_-_16870704 | 0.06 |
ENST00000380672.4 ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2 |
basonuclin 2 |
| chr11_+_63606558 | 0.06 |
ENST00000350490.7 ENST00000502399.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr17_-_39507064 | 0.06 |
ENST00000007735.3 |
KRT33A |
keratin 33A |
| chr4_-_121993673 | 0.06 |
ENST00000379692.4 |
NDNF |
neuron-derived neurotrophic factor |
| chr4_-_66536196 | 0.06 |
ENST00000511294.1 |
EPHA5 |
EPH receptor A5 |
| chr15_-_71407833 | 0.06 |
ENST00000449977.2 |
CT62 |
cancer/testis antigen 62 |
| chr19_+_10563567 | 0.05 |
ENST00000344979.3 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr1_+_16348497 | 0.05 |
ENST00000439316.2 |
CLCNKA |
chloride channel, voltage-sensitive Ka |
| chr1_+_1567474 | 0.05 |
ENST00000356026.5 |
MMP23B |
matrix metallopeptidase 23B |
| chr10_+_50818343 | 0.05 |
ENST00000374115.3 |
SLC18A3 |
solute carrier family 18 (vesicular acetylcholine transporter), member 3 |
| chr19_-_46142680 | 0.05 |
ENST00000245925.3 |
EML2 |
echinoderm microtubule associated protein like 2 |
| chr19_-_42931567 | 0.04 |
ENST00000244289.4 |
LIPE |
lipase, hormone-sensitive |
| chr7_+_39017504 | 0.04 |
ENST00000403058.1 |
POU6F2 |
POU class 6 homeobox 2 |
| chr22_+_21369316 | 0.04 |
ENST00000413302.2 ENST00000402329.3 ENST00000336296.2 ENST00000401443.1 ENST00000443995.3 |
P2RX6 |
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr11_+_63606477 | 0.04 |
ENST00000508192.1 ENST00000361128.5 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr17_-_18945798 | 0.03 |
ENST00000395635.1 |
GRAP |
GRB2-related adaptor protein |
| chr11_-_22647350 | 0.03 |
ENST00000327470.3 |
FANCF |
Fanconi anemia, complementation group F |
| chrX_+_18725758 | 0.03 |
ENST00000472826.1 ENST00000544635.1 ENST00000496075.2 |
PPEF1 |
protein phosphatase, EF-hand calcium binding domain 1 |
| chr6_-_87804815 | 0.03 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr11_+_15095108 | 0.03 |
ENST00000324229.6 ENST00000533448.1 |
CALCB |
calcitonin-related polypeptide beta |
| chr3_-_45837959 | 0.03 |
ENST00000353278.4 ENST00000456124.2 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr14_+_22739823 | 0.03 |
ENST00000390464.2 |
TRAV38-1 |
T cell receptor alpha variable 38-1 |
| chr19_-_46142637 | 0.03 |
ENST00000590043.1 ENST00000589876.1 |
EML2 |
echinoderm microtubule associated protein like 2 |
| chr10_+_118350468 | 0.03 |
ENST00000358834.4 ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1 |
pancreatic lipase-related protein 1 |
| chr15_-_71407806 | 0.03 |
ENST00000566432.1 ENST00000567117.1 |
CT62 |
cancer/testis antigen 62 |
| chr1_+_44870866 | 0.02 |
ENST00000355387.2 ENST00000361799.2 |
RNF220 |
ring finger protein 220 |
| chr1_+_22962948 | 0.02 |
ENST00000374642.3 |
C1QA |
complement component 1, q subcomponent, A chain |
| chr11_+_63606373 | 0.02 |
ENST00000402010.2 ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr2_-_89399845 | 0.02 |
ENST00000479981.1 |
IGKV1-16 |
immunoglobulin kappa variable 1-16 |
| chr22_+_41956767 | 0.02 |
ENST00000306149.7 |
CSDC2 |
cold shock domain containing C2, RNA binding |
| chr12_-_11002063 | 0.02 |
ENST00000544994.1 ENST00000228811.4 ENST00000540107.1 |
PRR4 |
proline rich 4 (lacrimal) |
| chr1_+_17559776 | 0.02 |
ENST00000537499.1 ENST00000413717.2 ENST00000536552.1 |
PADI1 |
peptidyl arginine deiminase, type I |
| chrX_+_91034260 | 0.02 |
ENST00000395337.2 |
PCDH11X |
protocadherin 11 X-linked |
| chr10_+_60094735 | 0.02 |
ENST00000373910.4 |
UBE2D1 |
ubiquitin-conjugating enzyme E2D 1 |
| chr17_-_39191107 | 0.02 |
ENST00000344363.5 |
KRTAP1-3 |
keratin associated protein 1-3 |
| chr4_+_111397216 | 0.01 |
ENST00000265162.5 |
ENPEP |
glutamyl aminopeptidase (aminopeptidase A) |
| chr6_+_31730773 | 0.01 |
ENST00000415669.2 ENST00000425424.1 |
SAPCD1 |
suppressor APC domain containing 1 |
| chr11_-_2182388 | 0.01 |
ENST00000421783.1 ENST00000397262.1 ENST00000250971.3 ENST00000381330.4 ENST00000397270.1 |
INS INS-IGF2 |
insulin INS-IGF2 readthrough |
| chr4_+_95917383 | 0.01 |
ENST00000512312.1 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr2_+_89184868 | 0.01 |
ENST00000390243.2 |
IGKV4-1 |
immunoglobulin kappa variable 4-1 |
| chrX_+_53078273 | 0.01 |
ENST00000332582.4 |
GPR173 |
G protein-coupled receptor 173 |
| chr2_-_99552620 | 0.01 |
ENST00000428096.1 ENST00000397899.2 ENST00000420294.1 |
KIAA1211L |
KIAA1211-like |
| chr3_-_45838011 | 0.01 |
ENST00000358525.4 ENST00000413781.1 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr4_-_66535653 | 0.01 |
ENST00000354839.4 ENST00000432638.2 |
EPHA5 |
EPH receptor A5 |
| chr12_-_11036844 | 0.01 |
ENST00000428168.2 |
PRH1 |
proline-rich protein HaeIII subfamily 1 |
| chr17_-_56609302 | 0.00 |
ENST00000581607.1 ENST00000317256.6 ENST00000426861.1 ENST00000580809.1 ENST00000577729.1 ENST00000583291.1 |
SEPT4 |
septin 4 |
| chr2_+_11679963 | 0.00 |
ENST00000263834.5 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr2_+_90248739 | 0.00 |
ENST00000468879.1 |
IGKV1D-43 |
immunoglobulin kappa variable 1D-43 |
| chr15_+_80351910 | 0.00 |
ENST00000261749.6 ENST00000561060.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 2.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.9 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.2 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.5 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.4 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.5 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 0.0 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.9 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.0 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.2 | 0.9 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.2 | 0.6 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.4 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.5 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 0.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.4 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.6 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.5 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.7 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.3 | GO:0090296 | regulation of mitochondrial DNA replication(GO:0090296) |
| 0.1 | 0.4 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.3 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.3 | GO:1904044 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) response to aldosterone(GO:1904044) |
| 0.0 | 0.2 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.2 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.0 | 0.4 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.6 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.2 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.2 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:0038170 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.1 | GO:2000969 | positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.9 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.7 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.4 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.2 | GO:0060340 | MDA-5 signaling pathway(GO:0039530) positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.0 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.0 | 0.2 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0038202 | TORC1 signaling(GO:0038202) |