Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
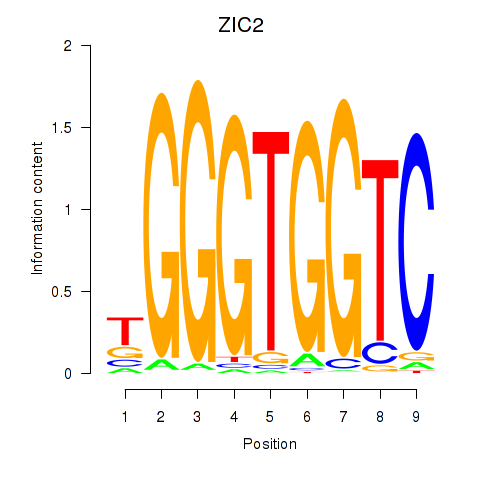
Results for ZIC2_GLI1
Z-value: 0.69
Transcription factors associated with ZIC2_GLI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC2
|
ENSG00000043355.6 | ZIC2 |
|
GLI1
|
ENSG00000111087.5 | GLI1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLI1 | hg19_v2_chr12_+_57853918_57853934 | 0.32 | 2.3e-01 | Click! |
Activity profile of ZIC2_GLI1 motif
Sorted Z-values of ZIC2_GLI1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC2_GLI1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_69064300 | 1.24 |
ENST00000342771.4 |
AUTS2 |
autism susceptibility candidate 2 |
| chr11_-_111783595 | 1.23 |
ENST00000528628.1 |
CRYAB |
crystallin, alpha B |
| chr11_-_111783919 | 1.21 |
ENST00000531198.1 ENST00000533879.1 |
CRYAB |
crystallin, alpha B |
| chr7_+_130131907 | 0.99 |
ENST00000223215.4 ENST00000437945.1 |
MEST |
mesoderm specific transcript |
| chr11_+_111782934 | 0.95 |
ENST00000304298.3 |
HSPB2 |
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr7_+_119913688 | 0.92 |
ENST00000331113.4 |
KCND2 |
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr19_-_44285401 | 0.89 |
ENST00000262888.3 |
KCNN4 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr1_+_209941827 | 0.87 |
ENST00000367023.1 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
| chr10_-_90712520 | 0.84 |
ENST00000224784.6 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr11_+_111783450 | 0.78 |
ENST00000537382.1 |
HSPB2 |
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr17_-_40835076 | 0.76 |
ENST00000591765.1 |
CCR10 |
chemokine (C-C motif) receptor 10 |
| chr20_+_57430162 | 0.74 |
ENST00000450130.1 ENST00000349036.3 ENST00000423897.1 |
GNAS |
GNAS complex locus |
| chr1_+_209941942 | 0.64 |
ENST00000487271.1 ENST00000477431.1 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
| chr6_-_29595779 | 0.59 |
ENST00000355973.3 ENST00000377012.4 |
GABBR1 |
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr3_-_72496035 | 0.47 |
ENST00000477973.2 |
RYBP |
RING1 and YY1 binding protein |
| chr10_-_95360983 | 0.43 |
ENST00000371464.3 |
RBP4 |
retinol binding protein 4, plasma |
| chr3_+_133118839 | 0.43 |
ENST00000302334.2 |
BFSP2 |
beaded filament structural protein 2, phakinin |
| chr11_-_111784005 | 0.39 |
ENST00000527899.1 |
CRYAB |
crystallin, alpha B |
| chr12_+_72148614 | 0.38 |
ENST00000261263.3 |
RAB21 |
RAB21, member RAS oncogene family |
| chr7_-_92465868 | 0.37 |
ENST00000424848.2 |
CDK6 |
cyclin-dependent kinase 6 |
| chr14_-_64971288 | 0.37 |
ENST00000394715.1 |
ZBTB25 |
zinc finger and BTB domain containing 25 |
| chr19_+_676385 | 0.36 |
ENST00000166139.4 |
FSTL3 |
follistatin-like 3 (secreted glycoprotein) |
| chr16_+_30675654 | 0.34 |
ENST00000287468.5 ENST00000395073.2 |
FBRS |
fibrosin |
| chr2_+_46769798 | 0.34 |
ENST00000238738.4 |
RHOQ |
ras homolog family member Q |
| chr20_+_30640004 | 0.34 |
ENST00000520553.1 ENST00000518730.1 ENST00000375852.2 |
HCK |
hemopoietic cell kinase |
| chr20_+_30639991 | 0.33 |
ENST00000534862.1 ENST00000538448.1 ENST00000375862.2 |
HCK |
hemopoietic cell kinase |
| chr9_-_98279241 | 0.32 |
ENST00000437951.1 ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1 |
patched 1 |
| chr3_-_52486841 | 0.32 |
ENST00000496590.1 |
TNNC1 |
troponin C type 1 (slow) |
| chr5_-_150460539 | 0.31 |
ENST00000520931.1 ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr11_-_111782484 | 0.31 |
ENST00000533971.1 |
CRYAB |
crystallin, alpha B |
| chr11_-_111782696 | 0.30 |
ENST00000227251.3 ENST00000526180.1 |
CRYAB |
crystallin, alpha B |
| chr14_+_24583836 | 0.30 |
ENST00000559115.1 ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
| chr5_+_157158205 | 0.29 |
ENST00000231198.7 |
THG1L |
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr14_+_24584508 | 0.29 |
ENST00000559354.1 ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
| chr6_+_43739697 | 0.28 |
ENST00000230480.6 |
VEGFA |
vascular endothelial growth factor A |
| chr15_+_81591757 | 0.28 |
ENST00000558332.1 |
IL16 |
interleukin 16 |
| chr12_+_57522258 | 0.27 |
ENST00000553277.1 ENST00000243077.3 |
LRP1 |
low density lipoprotein receptor-related protein 1 |
| chrX_+_51636629 | 0.27 |
ENST00000375722.1 ENST00000326587.7 ENST00000375695.2 |
MAGED1 |
melanoma antigen family D, 1 |
| chr6_-_32784687 | 0.27 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr16_+_19183671 | 0.26 |
ENST00000562711.2 |
SYT17 |
synaptotagmin XVII |
| chr17_+_40834580 | 0.25 |
ENST00000264638.4 |
CNTNAP1 |
contactin associated protein 1 |
| chr6_-_43337180 | 0.25 |
ENST00000318149.3 ENST00000361428.2 |
ZNF318 |
zinc finger protein 318 |
| chr6_-_32821599 | 0.25 |
ENST00000354258.4 |
TAP1 |
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr2_-_28113965 | 0.25 |
ENST00000302188.3 |
RBKS |
ribokinase |
| chr8_-_117886955 | 0.25 |
ENST00000297338.2 |
RAD21 |
RAD21 homolog (S. pombe) |
| chr19_+_4969116 | 0.25 |
ENST00000588337.1 ENST00000159111.4 ENST00000381759.4 |
KDM4B |
lysine (K)-specific demethylase 4B |
| chr5_-_150460914 | 0.24 |
ENST00000389378.2 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr3_-_73673991 | 0.23 |
ENST00000308537.4 ENST00000263666.4 |
PDZRN3 |
PDZ domain containing ring finger 3 |
| chr20_+_43343517 | 0.21 |
ENST00000372865.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr14_-_61124977 | 0.21 |
ENST00000554986.1 |
SIX1 |
SIX homeobox 1 |
| chr4_+_77870960 | 0.20 |
ENST00000505788.1 ENST00000510515.1 ENST00000504637.1 |
SEPT11 |
septin 11 |
| chr14_+_64971292 | 0.19 |
ENST00000358738.3 ENST00000394712.2 |
ZBTB1 |
zinc finger and BTB domain containing 1 |
| chr20_+_43343476 | 0.19 |
ENST00000372868.2 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr5_+_161275320 | 0.19 |
ENST00000437025.2 |
GABRA1 |
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr15_-_78526855 | 0.19 |
ENST00000541759.1 ENST00000558130.1 |
ACSBG1 |
acyl-CoA synthetase bubblegum family member 1 |
| chr13_+_20532807 | 0.18 |
ENST00000382869.3 ENST00000382881.3 |
ZMYM2 |
zinc finger, MYM-type 2 |
| chr12_-_122985494 | 0.18 |
ENST00000336229.4 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
| chr12_-_122985067 | 0.17 |
ENST00000540586.1 ENST00000543897.1 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
| chr12_-_6798616 | 0.17 |
ENST00000355772.4 ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384 |
zinc finger protein 384 |
| chr10_-_97050777 | 0.17 |
ENST00000329399.6 |
PDLIM1 |
PDZ and LIM domain 1 |
| chr1_-_182573514 | 0.17 |
ENST00000367558.5 |
RGS16 |
regulator of G-protein signaling 16 |
| chr19_-_55881741 | 0.17 |
ENST00000264563.2 ENST00000590625.1 ENST00000585513.1 |
IL11 |
interleukin 11 |
| chr20_+_43343886 | 0.16 |
ENST00000190983.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr4_+_77870856 | 0.16 |
ENST00000264893.6 ENST00000502584.1 ENST00000510641.1 |
SEPT11 |
septin 11 |
| chr12_-_6798410 | 0.16 |
ENST00000361959.3 ENST00000436774.2 ENST00000544482.1 |
ZNF384 |
zinc finger protein 384 |
| chr6_+_31620191 | 0.15 |
ENST00000375918.2 ENST00000375920.4 |
APOM |
apolipoprotein M |
| chr1_-_147142557 | 0.15 |
ENST00000369238.6 |
ACP6 |
acid phosphatase 6, lysophosphatidic |
| chr19_-_13617037 | 0.14 |
ENST00000360228.5 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
| chr12_-_6798523 | 0.14 |
ENST00000319770.3 |
ZNF384 |
zinc finger protein 384 |
| chr17_+_38497640 | 0.14 |
ENST00000394086.3 |
RARA |
retinoic acid receptor, alpha |
| chr17_+_43299156 | 0.13 |
ENST00000331495.3 |
FMNL1 |
formin-like 1 |
| chr5_+_52776449 | 0.13 |
ENST00000396947.3 |
FST |
follistatin |
| chr3_-_8686479 | 0.13 |
ENST00000544814.1 ENST00000427408.1 |
SSUH2 |
ssu-2 homolog (C. elegans) |
| chr17_-_7137582 | 0.12 |
ENST00000575756.1 ENST00000575458.1 |
DVL2 |
dishevelled segment polarity protein 2 |
| chr13_+_111767650 | 0.12 |
ENST00000449979.1 ENST00000370623.3 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
| chrX_+_12809463 | 0.12 |
ENST00000380663.3 ENST00000380668.5 ENST00000398491.2 ENST00000489404.1 |
PRPS2 |
phosphoribosyl pyrophosphate synthetase 2 |
| chr19_-_39330818 | 0.12 |
ENST00000594769.1 ENST00000602021.1 |
AC104534.3 |
Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial |
| chr9_+_131174024 | 0.12 |
ENST00000420034.1 ENST00000372842.1 |
CERCAM |
cerebral endothelial cell adhesion molecule |
| chr11_+_117049910 | 0.12 |
ENST00000431081.2 ENST00000524842.1 |
SIDT2 |
SID1 transmembrane family, member 2 |
| chr1_+_154966058 | 0.12 |
ENST00000392487.1 |
LENEP |
lens epithelial protein |
| chr12_-_57522813 | 0.11 |
ENST00000556155.1 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr2_+_97481974 | 0.11 |
ENST00000377060.3 ENST00000305510.3 |
CNNM3 |
cyclin M3 |
| chr3_+_4535025 | 0.11 |
ENST00000302640.8 ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr7_-_91875109 | 0.11 |
ENST00000412043.2 ENST00000430102.1 ENST00000425073.1 ENST00000394503.2 ENST00000454017.1 ENST00000440209.1 ENST00000413688.1 ENST00000452773.1 ENST00000433016.1 ENST00000394505.2 ENST00000422347.1 ENST00000458493.1 ENST00000425919.1 |
KRIT1 |
KRIT1, ankyrin repeat containing |
| chr15_-_26108355 | 0.11 |
ENST00000356865.6 |
ATP10A |
ATPase, class V, type 10A |
| chrX_+_47444613 | 0.11 |
ENST00000445623.1 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
| chr9_+_8858102 | 0.11 |
ENST00000447950.1 ENST00000430766.1 |
RP11-75C9.1 |
RP11-75C9.1 |
| chr16_+_75256507 | 0.11 |
ENST00000495583.1 |
CTRB1 |
chymotrypsinogen B1 |
| chr9_+_34989638 | 0.10 |
ENST00000453597.3 ENST00000335998.3 ENST00000312316.5 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr17_-_76778339 | 0.10 |
ENST00000591455.1 ENST00000446868.3 ENST00000361101.4 ENST00000589296.1 |
CYTH1 |
cytohesin 1 |
| chr18_-_12884259 | 0.10 |
ENST00000353319.4 ENST00000327283.3 |
PTPN2 |
protein tyrosine phosphatase, non-receptor type 2 |
| chr9_-_115818951 | 0.10 |
ENST00000553380.1 ENST00000374227.3 |
ZFP37 |
ZFP37 zinc finger protein |
| chr6_+_26240561 | 0.10 |
ENST00000377745.2 |
HIST1H4F |
histone cluster 1, H4f |
| chr12_-_49333446 | 0.09 |
ENST00000537495.1 |
AC073610.5 |
Uncharacterized protein |
| chrX_+_70443050 | 0.09 |
ENST00000361726.6 |
GJB1 |
gap junction protein, beta 1, 32kDa |
| chr1_+_36690011 | 0.09 |
ENST00000354618.5 ENST00000469141.2 ENST00000478853.1 |
THRAP3 |
thyroid hormone receptor associated protein 3 |
| chr9_+_34990219 | 0.09 |
ENST00000541010.1 ENST00000454002.2 ENST00000545841.1 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr1_-_150208498 | 0.08 |
ENST00000314136.8 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr3_+_49449636 | 0.08 |
ENST00000273590.3 |
TCTA |
T-cell leukemia translocation altered |
| chr8_+_98656693 | 0.08 |
ENST00000519934.1 |
MTDH |
metadherin |
| chr17_-_27278304 | 0.08 |
ENST00000577226.1 |
PHF12 |
PHD finger protein 12 |
| chr2_+_106361333 | 0.08 |
ENST00000233154.4 ENST00000451463.2 |
NCK2 |
NCK adaptor protein 2 |
| chr4_+_72052964 | 0.08 |
ENST00000264485.5 ENST00000425175.1 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr15_-_23891175 | 0.08 |
ENST00000532292.1 |
MAGEL2 |
MAGE-like 2 |
| chr8_-_71581377 | 0.08 |
ENST00000276590.4 ENST00000522447.1 |
LACTB2 |
lactamase, beta 2 |
| chr5_+_162864575 | 0.07 |
ENST00000512163.1 ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1 |
cyclin G1 |
| chr11_+_34938119 | 0.07 |
ENST00000227868.4 ENST00000430469.2 ENST00000533262.1 |
PDHX |
pyruvate dehydrogenase complex, component X |
| chr4_-_176733897 | 0.07 |
ENST00000393658.2 |
GPM6A |
glycoprotein M6A |
| chr18_-_31803435 | 0.07 |
ENST00000589544.1 ENST00000269185.4 ENST00000261592.5 |
NOL4 |
nucleolar protein 4 |
| chr22_+_38035459 | 0.07 |
ENST00000357436.4 |
SH3BP1 |
SH3-domain binding protein 1 |
| chr9_+_33264861 | 0.07 |
ENST00000223500.8 |
CHMP5 |
charged multivesicular body protein 5 |
| chr14_-_88789581 | 0.07 |
ENST00000319231.5 |
KCNK10 |
potassium channel, subfamily K, member 10 |
| chr9_+_33265011 | 0.07 |
ENST00000419016.2 |
CHMP5 |
charged multivesicular body protein 5 |
| chr9_+_136399929 | 0.07 |
ENST00000393060.1 |
ADAMTSL2 |
ADAMTS-like 2 |
| chr4_-_186697044 | 0.07 |
ENST00000437304.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr17_+_17380294 | 0.07 |
ENST00000268711.3 ENST00000580462.1 |
MED9 |
mediator complex subunit 9 |
| chr12_+_31477250 | 0.07 |
ENST00000313737.4 |
AC024940.1 |
AC024940.1 |
| chr11_+_10477733 | 0.07 |
ENST00000528723.1 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr20_+_57427765 | 0.07 |
ENST00000371100.4 |
GNAS |
GNAS complex locus |
| chr1_-_150208412 | 0.07 |
ENST00000532744.1 ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr17_+_4843679 | 0.06 |
ENST00000576229.1 |
RNF167 |
ring finger protein 167 |
| chr2_-_208989225 | 0.06 |
ENST00000264376.4 |
CRYGD |
crystallin, gamma D |
| chr19_+_36208877 | 0.06 |
ENST00000420124.1 ENST00000222270.7 ENST00000341701.1 |
KMT2B |
Histone-lysine N-methyltransferase 2B |
| chr16_-_16317321 | 0.06 |
ENST00000205557.7 ENST00000575728.1 |
ABCC6 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
| chr14_+_92980111 | 0.06 |
ENST00000216487.7 ENST00000557762.1 |
RIN3 |
Ras and Rab interactor 3 |
| chr17_-_7137857 | 0.06 |
ENST00000005340.5 |
DVL2 |
dishevelled segment polarity protein 2 |
| chr11_-_46142615 | 0.06 |
ENST00000529734.1 ENST00000323180.6 |
PHF21A |
PHD finger protein 21A |
| chr11_+_34073269 | 0.06 |
ENST00000389645.3 |
CAPRIN1 |
cell cycle associated protein 1 |
| chr3_-_128902759 | 0.06 |
ENST00000422453.2 ENST00000504813.1 ENST00000512338.1 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
| chr11_+_34073195 | 0.06 |
ENST00000341394.4 |
CAPRIN1 |
cell cycle associated protein 1 |
| chr17_+_73997796 | 0.06 |
ENST00000586261.1 |
CDK3 |
cyclin-dependent kinase 3 |
| chr19_+_45312347 | 0.05 |
ENST00000270233.6 ENST00000591520.1 |
BCAM |
basal cell adhesion molecule (Lutheran blood group) |
| chr2_+_136289030 | 0.05 |
ENST00000409478.1 ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1 |
R3H domain containing 1 |
| chr1_-_201081579 | 0.05 |
ENST00000367338.3 ENST00000362061.3 |
CACNA1S |
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr2_+_28113583 | 0.05 |
ENST00000344773.2 ENST00000379624.1 ENST00000342045.2 ENST00000379632.2 ENST00000361704.2 |
BRE |
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr5_+_52776228 | 0.05 |
ENST00000256759.3 |
FST |
follistatin |
| chr3_-_49449521 | 0.05 |
ENST00000431929.1 ENST00000418115.1 |
RHOA |
ras homolog family member A |
| chr3_-_128902729 | 0.05 |
ENST00000451728.2 ENST00000446936.2 ENST00000502976.1 ENST00000500450.2 ENST00000441626.2 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
| chr9_-_33264557 | 0.05 |
ENST00000473781.1 ENST00000488499.1 |
BAG1 |
BCL2-associated athanogene |
| chr6_+_33172407 | 0.05 |
ENST00000374662.3 |
HSD17B8 |
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr7_-_92463210 | 0.05 |
ENST00000265734.4 |
CDK6 |
cyclin-dependent kinase 6 |
| chr1_-_115053781 | 0.04 |
ENST00000358465.2 ENST00000369543.2 |
TRIM33 |
tripartite motif containing 33 |
| chr11_-_30607819 | 0.04 |
ENST00000448418.2 |
MPPED2 |
metallophosphoesterase domain containing 2 |
| chr15_+_101142722 | 0.04 |
ENST00000332783.7 ENST00000558747.1 ENST00000343276.4 |
ASB7 |
ankyrin repeat and SOCS box containing 7 |
| chr14_-_23834411 | 0.04 |
ENST00000429593.2 |
EFS |
embryonal Fyn-associated substrate |
| chr1_-_45308616 | 0.04 |
ENST00000447098.2 ENST00000372192.3 |
PTCH2 |
patched 2 |
| chr10_-_48416849 | 0.04 |
ENST00000249598.1 |
GDF2 |
growth differentiation factor 2 |
| chr2_-_68694390 | 0.04 |
ENST00000377957.3 |
FBXO48 |
F-box protein 48 |
| chr17_+_4843654 | 0.04 |
ENST00000575111.1 |
RNF167 |
ring finger protein 167 |
| chr9_-_33264676 | 0.04 |
ENST00000472232.3 ENST00000379704.2 |
BAG1 |
BCL2-associated athanogene |
| chr3_+_110790590 | 0.04 |
ENST00000485303.1 |
PVRL3 |
poliovirus receptor-related 3 |
| chr5_-_89705537 | 0.04 |
ENST00000522864.1 ENST00000522083.1 ENST00000522565.1 ENST00000522842.1 ENST00000283122.3 |
CETN3 |
centrin, EF-hand protein, 3 |
| chr4_+_62067860 | 0.04 |
ENST00000514591.1 |
LPHN3 |
latrophilin 3 |
| chr10_+_180405 | 0.04 |
ENST00000439456.1 ENST00000397962.3 ENST00000309776.4 ENST00000381602.4 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
| chr20_-_34025999 | 0.04 |
ENST00000374369.3 |
GDF5 |
growth differentiation factor 5 |
| chr11_-_34938039 | 0.04 |
ENST00000395787.3 |
APIP |
APAF1 interacting protein |
| chr16_-_68002456 | 0.04 |
ENST00000576616.1 ENST00000572037.1 ENST00000338335.3 ENST00000422611.2 ENST00000316341.3 |
SLC12A4 |
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr3_+_4535155 | 0.04 |
ENST00000544951.1 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr5_+_175792459 | 0.04 |
ENST00000310389.5 |
ARL10 |
ADP-ribosylation factor-like 10 |
| chr9_-_115819039 | 0.04 |
ENST00000555206.1 |
ZFP37 |
ZFP37 zinc finger protein |
| chr12_+_57853918 | 0.03 |
ENST00000532291.1 ENST00000543426.1 ENST00000228682.2 ENST00000546141.1 |
GLI1 |
GLI family zinc finger 1 |
| chr11_+_33278811 | 0.03 |
ENST00000303296.4 ENST00000379016.3 |
HIPK3 |
homeodomain interacting protein kinase 3 |
| chr13_+_20532900 | 0.03 |
ENST00000382871.2 |
ZMYM2 |
zinc finger, MYM-type 2 |
| chr4_-_186696425 | 0.03 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr1_-_111217603 | 0.03 |
ENST00000369769.2 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
| chr13_+_20532848 | 0.03 |
ENST00000382874.2 |
ZMYM2 |
zinc finger, MYM-type 2 |
| chr19_-_6424783 | 0.03 |
ENST00000398148.3 |
KHSRP |
KH-type splicing regulatory protein |
| chr19_+_58790314 | 0.03 |
ENST00000196548.5 ENST00000608843.1 |
ZNF8 ZNF8 |
Zinc finger protein 8 zinc finger protein 8 |
| chr17_+_26989109 | 0.03 |
ENST00000314616.6 ENST00000347486.4 |
SUPT6H |
suppressor of Ty 6 homolog (S. cerevisiae) |
| chr11_+_64001962 | 0.03 |
ENST00000309422.2 |
VEGFB |
vascular endothelial growth factor B |
| chr16_+_28834303 | 0.03 |
ENST00000340394.8 ENST00000325215.6 ENST00000395547.2 ENST00000336783.4 ENST00000382686.4 ENST00000564304.1 |
ATXN2L |
ataxin 2-like |
| chr8_+_98656336 | 0.03 |
ENST00000336273.3 |
MTDH |
metadherin |
| chr22_-_46373004 | 0.03 |
ENST00000339464.4 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
| chr14_+_22984601 | 0.03 |
ENST00000390509.1 |
TRAJ28 |
T cell receptor alpha joining 28 |
| chr15_-_81282133 | 0.02 |
ENST00000261758.4 |
MESDC2 |
mesoderm development candidate 2 |
| chr1_-_150208363 | 0.02 |
ENST00000436748.2 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr15_-_78526942 | 0.02 |
ENST00000258873.4 |
ACSBG1 |
acyl-CoA synthetase bubblegum family member 1 |
| chr17_-_40540484 | 0.02 |
ENST00000588969.1 |
STAT3 |
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr2_-_51259641 | 0.02 |
ENST00000406316.2 ENST00000405581.1 |
NRXN1 |
neurexin 1 |
| chr6_-_26027480 | 0.02 |
ENST00000377364.3 |
HIST1H4B |
histone cluster 1, H4b |
| chr20_+_3451650 | 0.02 |
ENST00000262919.5 |
ATRN |
attractin |
| chr19_-_40919271 | 0.02 |
ENST00000291825.7 ENST00000324001.7 |
PRX |
periaxin |
| chr9_-_35958151 | 0.02 |
ENST00000341959.2 |
OR2S2 |
olfactory receptor, family 2, subfamily S, member 2 |
| chr21_-_36259445 | 0.02 |
ENST00000399240.1 |
RUNX1 |
runt-related transcription factor 1 |
| chr9_-_122131696 | 0.01 |
ENST00000373964.2 ENST00000265922.3 |
BRINP1 |
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
| chr11_+_64009072 | 0.01 |
ENST00000535135.1 ENST00000394540.3 |
FKBP2 |
FK506 binding protein 2, 13kDa |
| chr10_-_35104185 | 0.01 |
ENST00000374789.3 ENST00000374788.3 ENST00000346874.4 ENST00000374794.3 ENST00000350537.4 ENST00000374790.3 ENST00000374776.1 ENST00000374773.1 ENST00000545693.1 ENST00000545260.1 ENST00000340077.5 |
PARD3 |
par-3 family cell polarity regulator |
| chr5_-_168727786 | 0.01 |
ENST00000332966.8 |
SLIT3 |
slit homolog 3 (Drosophila) |
| chr5_-_148758839 | 0.01 |
ENST00000261796.3 |
IL17B |
interleukin 17B |
| chr7_-_99679324 | 0.01 |
ENST00000292393.5 ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3 |
zinc finger protein 3 |
| chr10_-_98346801 | 0.01 |
ENST00000371142.4 |
TM9SF3 |
transmembrane 9 superfamily member 3 |
| chr11_-_46142948 | 0.01 |
ENST00000257821.4 |
PHF21A |
PHD finger protein 21A |
| chr3_+_137728842 | 0.01 |
ENST00000183605.5 |
CLDN18 |
claudin 18 |
| chr18_-_31803169 | 0.01 |
ENST00000590712.1 |
NOL4 |
nucleolar protein 4 |
| chr4_-_38666430 | 0.01 |
ENST00000436901.1 |
AC021860.1 |
Uncharacterized protein |
| chr1_+_24286287 | 0.01 |
ENST00000334351.7 ENST00000374468.1 |
PNRC2 |
proline-rich nuclear receptor coactivator 2 |
| chr1_+_226736446 | 0.01 |
ENST00000366788.3 ENST00000366789.4 |
C1orf95 |
chromosome 1 open reading frame 95 |
| chr2_+_25015968 | 0.01 |
ENST00000380834.2 ENST00000473706.1 |
CENPO |
centromere protein O |
| chr11_-_30608413 | 0.01 |
ENST00000528686.1 |
MPPED2 |
metallophosphoesterase domain containing 2 |
| chr22_+_37447771 | 0.01 |
ENST00000402077.3 ENST00000403888.3 ENST00000456470.1 |
KCTD17 |
potassium channel tetramerization domain containing 17 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.2 | 0.7 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.2 | 3.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 0.5 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.8 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.8 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.3 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.3 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.1 | 1.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.4 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.2 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 0.3 | GO:0021997 | response to chlorate(GO:0010157) neural plate axis specification(GO:0021997) |
| 0.1 | 0.3 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.2 | GO:0061055 | myotome development(GO:0061055) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.5 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.1 | 0.2 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.2 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.4 | GO:1904628 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 0.9 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.3 | GO:0002584 | negative regulation of antigen processing and presentation of peptide antigen(GO:0002584) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.0 | 0.2 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.0 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.4 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.2 | GO:0034445 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:1903973 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.2 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.5 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.2 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.5 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.0 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.0 | 0.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.0 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.3 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.0 | GO:0021938 | ventral midline development(GO:0007418) smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.1 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.5 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.4 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 0.8 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.2 | 0.5 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 0.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.1 | 0.2 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.4 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 0.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.4 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 3.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 0.3 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.8 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.4 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.3 | GO:0042954 | apolipoprotein receptor activity(GO:0030226) lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.8 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.5 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.4 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.3 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.0 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |