Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
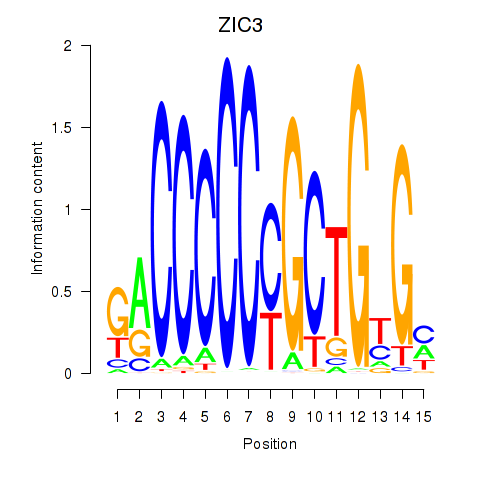
Results for ZIC3_ZIC4
Z-value: 1.10
Transcription factors associated with ZIC3_ZIC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC3
|
ENSG00000156925.7 | ZIC3 |
|
ZIC4
|
ENSG00000174963.13 | ZIC4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZIC3 | hg19_v2_chrX_+_136648297_136648319 | 0.14 | 6.2e-01 | Click! |
Activity profile of ZIC3_ZIC4 motif
Sorted Z-values of ZIC3_ZIC4 motif
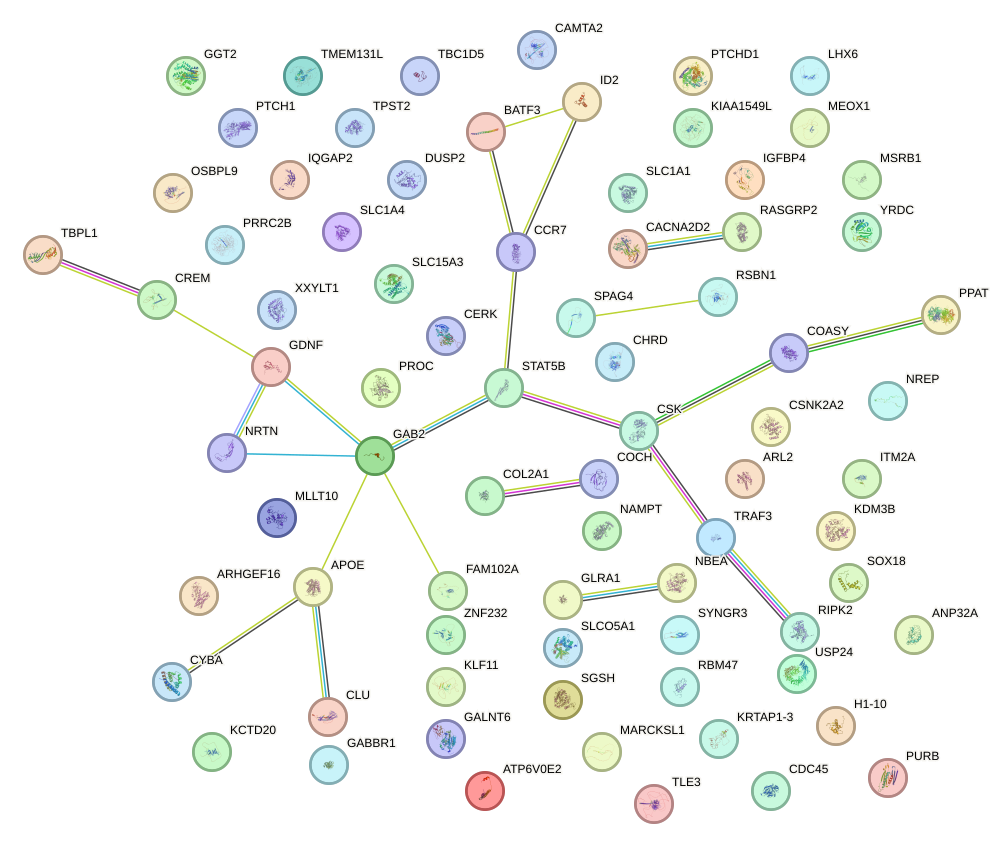
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC3_ZIC4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_32801825 | 3.68 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chr14_+_31343747 | 2.59 |
ENST00000216361.4 ENST00000396618.3 ENST00000475087.1 |
COCH |
cochlin |
| chr5_+_75699149 | 2.54 |
ENST00000379730.3 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr16_-_88717482 | 2.47 |
ENST00000261623.3 |
CYBA |
cytochrome b-245, alpha polypeptide |
| chr5_+_75699040 | 2.36 |
ENST00000274364.6 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr20_+_34203794 | 2.13 |
ENST00000374273.3 |
SPAG4 |
sperm associated antigen 4 |
| chr4_-_40631859 | 1.76 |
ENST00000295971.7 ENST00000319592.4 |
RBM47 |
RNA binding motif protein 47 |
| chr20_+_35201857 | 1.32 |
ENST00000373874.2 |
TGIF2 |
TGFB-induced factor homeobox 2 |
| chr20_+_35201993 | 1.19 |
ENST00000373872.4 |
TGIF2 |
TGFB-induced factor homeobox 2 |
| chr9_-_130742792 | 1.08 |
ENST00000373095.1 |
FAM102A |
family with sequence similarity 102, member A |
| chrX_-_78622805 | 1.07 |
ENST00000373298.2 |
ITM2A |
integral membrane protein 2A |
| chr11_-_78128811 | 0.92 |
ENST00000530915.1 ENST00000361507.4 |
GAB2 |
GRB2-associated binding protein 2 |
| chr1_-_114355083 | 0.89 |
ENST00000261441.5 |
RSBN1 |
round spermatid basic protein 1 |
| chr7_+_149571045 | 0.88 |
ENST00000479613.1 ENST00000606024.1 ENST00000464662.1 ENST00000425642.2 |
ATP6V0E2 |
ATPase, H+ transporting V0 subunit e2 |
| chr7_-_994302 | 0.78 |
ENST00000265846.5 |
ADAP1 |
ArfGAP with dual PH domains 1 |
| chr5_+_121647877 | 0.78 |
ENST00000514497.2 ENST00000261367.7 |
SNCAIP |
synuclein, alpha interacting protein |
| chr17_-_38721711 | 0.77 |
ENST00000578085.1 ENST00000246657.2 |
CCR7 |
chemokine (C-C motif) receptor 7 |
| chr11_+_22688150 | 0.76 |
ENST00000454584.2 |
GAS2 |
growth arrest-specific 2 |
| chr5_+_121647764 | 0.74 |
ENST00000261368.8 ENST00000379533.2 ENST00000379536.2 ENST00000379538.3 |
SNCAIP |
synuclein, alpha interacting protein |
| chr16_-_58231782 | 0.72 |
ENST00000565188.1 ENST00000262506.3 |
CSNK2A2 |
casein kinase 2, alpha prime polypeptide |
| chr3_-_17783990 | 0.71 |
ENST00000429383.4 ENST00000446863.1 ENST00000414349.1 ENST00000428355.1 ENST00000425944.1 ENST00000445294.1 ENST00000444471.1 ENST00000415814.2 |
TBC1D5 |
TBC1 domain family, member 5 |
| chr12_-_48398104 | 0.70 |
ENST00000337299.6 ENST00000380518.3 |
COL2A1 |
collagen, type II, alpha 1 |
| chr15_+_75074385 | 0.69 |
ENST00000220003.9 |
CSK |
c-src tyrosine kinase |
| chr7_+_99775366 | 0.69 |
ENST00000394018.2 ENST00000416412.1 |
STAG3 |
stromal antigen 3 |
| chr15_+_75074410 | 0.69 |
ENST00000439220.2 |
CSK |
c-src tyrosine kinase |
| chr6_-_29595779 | 0.66 |
ENST00000355973.3 ENST00000377012.4 |
GABBR1 |
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr7_+_99775520 | 0.65 |
ENST00000317296.5 ENST00000422690.1 ENST00000439782.1 |
STAG3 |
stromal antigen 3 |
| chr10_+_21823079 | 0.61 |
ENST00000377100.3 ENST00000377072.3 ENST00000446906.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr10_+_35416223 | 0.60 |
ENST00000489321.1 ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM |
cAMP responsive element modulator |
| chr5_+_137688285 | 0.60 |
ENST00000314358.5 |
KDM3B |
lysine (K)-specific demethylase 3B |
| chr16_+_2039946 | 0.60 |
ENST00000248121.2 ENST00000568896.1 |
SYNGR3 |
synaptogyrin 3 |
| chr22_+_23134974 | 0.59 |
ENST00000390314.2 |
IGLV2-11 |
immunoglobulin lambda variable 2-11 |
| chr4_+_154387480 | 0.58 |
ENST00000409663.3 ENST00000440693.1 ENST00000409959.3 |
KIAA0922 |
KIAA0922 |
| chr13_+_35516390 | 0.58 |
ENST00000540320.1 ENST00000400445.3 ENST00000310336.4 |
NBEA |
neurobeachin |
| chr11_+_33563821 | 0.57 |
ENST00000321505.4 ENST00000265654.5 ENST00000389726.3 |
KIAA1549L |
KIAA1549-like |
| chr19_-_48753104 | 0.55 |
ENST00000447740.2 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr1_-_25256368 | 0.55 |
ENST00000308873.6 |
RUNX3 |
runt-related transcription factor 3 |
| chr5_+_121647924 | 0.54 |
ENST00000414317.2 |
SNCAIP |
synuclein, alpha interacting protein |
| chr11_-_73694346 | 0.54 |
ENST00000310473.3 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr17_-_40428359 | 0.54 |
ENST00000293328.3 |
STAT5B |
signal transducer and activator of transcription 5B |
| chr14_-_21994525 | 0.53 |
ENST00000538754.1 |
SALL2 |
spalt-like transcription factor 2 |
| chr19_-_48752812 | 0.53 |
ENST00000359009.4 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr7_-_105925558 | 0.53 |
ENST00000222553.3 |
NAMPT |
nicotinamide phosphoribosyltransferase |
| chr11_-_407103 | 0.52 |
ENST00000526395.1 |
SIGIRR |
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr22_-_20368028 | 0.50 |
ENST00000404912.1 |
GGTLC3 |
gamma-glutamyltransferase light chain 3 |
| chr2_+_8822113 | 0.50 |
ENST00000396290.1 ENST00000331129.3 |
ID2 |
inhibitor of DNA binding 2, dominant negative helix-loop-helix protein |
| chr1_-_212873267 | 0.48 |
ENST00000243440.1 |
BATF3 |
basic leucine zipper transcription factor, ATF-like 3 |
| chr7_+_150758304 | 0.46 |
ENST00000482950.1 ENST00000463414.1 ENST00000310317.5 |
SLC4A2 |
solute carrier family 4 (anion exchanger), member 2 |
| chr15_-_70388599 | 0.46 |
ENST00000560996.1 ENST00000558201.1 |
TLE3 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr22_-_47134077 | 0.46 |
ENST00000541677.1 ENST00000216264.8 |
CERK |
ceramide kinase |
| chr9_-_98279241 | 0.46 |
ENST00000437951.1 ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1 |
patched 1 |
| chr10_+_21823243 | 0.43 |
ENST00000307729.7 ENST00000377091.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr5_+_169064245 | 0.43 |
ENST00000256935.8 |
DOCK2 |
dedicator of cytokinesis 2 |
| chr3_-_129035120 | 0.43 |
ENST00000333762.4 |
H1FX |
H1 histone family, member X |
| chr2_+_128180842 | 0.42 |
ENST00000402125.2 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr5_-_151304337 | 0.42 |
ENST00000455880.2 ENST00000545569.1 ENST00000274576.4 |
GLRA1 |
glycine receptor, alpha 1 |
| chr20_-_62680984 | 0.42 |
ENST00000340356.7 |
SOX18 |
SRY (sex determining region Y)-box 18 |
| chr9_+_4490394 | 0.41 |
ENST00000262352.3 |
SLC1A1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| chr1_-_55680762 | 0.41 |
ENST00000407756.1 ENST00000294383.6 |
USP24 |
ubiquitin specific peptidase 24 |
| chr1_+_52195542 | 0.41 |
ENST00000462759.1 ENST00000486942.1 |
OSBPL9 |
oxysterol binding protein-like 9 |
| chr16_+_67596310 | 0.40 |
ENST00000264010.4 ENST00000401394.1 |
CTCF |
CCCTC-binding factor (zinc finger protein) |
| chr17_-_4890649 | 0.40 |
ENST00000361571.5 |
CAMTA2 |
calmodulin binding transcription activator 2 |
| chrX_-_151903184 | 0.39 |
ENST00000357916.4 ENST00000393869.3 |
MAGEA12 |
melanoma antigen family A, 12 |
| chr17_-_5026397 | 0.38 |
ENST00000250076.3 |
ZNF232 |
zinc finger protein 232 |
| chr12_-_51785182 | 0.38 |
ENST00000356317.3 ENST00000603188.1 ENST00000604847.1 ENST00000604506.1 |
GALNT6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
| chr1_-_38273840 | 0.38 |
ENST00000373044.2 |
YRDC |
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr5_-_111091948 | 0.38 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr17_+_38599693 | 0.38 |
ENST00000542955.1 ENST00000269593.4 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
| chr1_+_52195480 | 0.38 |
ENST00000531828.1 ENST00000361556.5 ENST00000481937.1 |
OSBPL9 |
oxysterol binding protein-like 9 |
| chr8_-_27462822 | 0.37 |
ENST00000522098.1 |
CLU |
clusterin |
| chr9_-_124989804 | 0.37 |
ENST00000373755.2 ENST00000373754.2 |
LHX6 |
LIM homeobox 6 |
| chr17_-_76124812 | 0.37 |
ENST00000592063.1 ENST00000589271.1 ENST00000322933.4 ENST00000589553.1 |
TMC6 |
transmembrane channel-like 6 |
| chr22_+_19467261 | 0.37 |
ENST00000455750.1 ENST00000437685.2 ENST00000263201.1 ENST00000404724.3 |
CDC45 |
cell division cycle 45 |
| chr19_+_45409011 | 0.36 |
ENST00000252486.4 ENST00000446996.1 ENST00000434152.1 |
APOE |
apolipoprotein E |
| chr15_-_69113218 | 0.36 |
ENST00000560303.1 ENST00000465139.2 |
ANP32A |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr4_-_57301748 | 0.36 |
ENST00000264220.2 |
PPAT |
phosphoribosyl pyrophosphate amidotransferase |
| chr16_+_30205754 | 0.35 |
ENST00000354723.6 ENST00000355544.5 |
SULT1A3 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr16_-_1993260 | 0.35 |
ENST00000361871.3 |
MSRB1 |
methionine sulfoxide reductase B1 |
| chr8_-_70745575 | 0.34 |
ENST00000524945.1 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr7_-_44924939 | 0.34 |
ENST00000395699.2 |
PURB |
purine-rich element binding protein B |
| chr4_-_140098339 | 0.33 |
ENST00000394235.2 |
ELF2 |
E74-like factor 2 (ets domain transcription factor) |
| chr2_-_96811170 | 0.33 |
ENST00000288943.4 |
DUSP2 |
dual specificity phosphatase 2 |
| chr17_-_39191107 | 0.33 |
ENST00000344363.5 |
KRTAP1-3 |
keratin associated protein 1-3 |
| chr8_+_90770008 | 0.32 |
ENST00000540020.1 |
RIPK2 |
receptor-interacting serine-threonine kinase 2 |
| chr6_+_134274322 | 0.32 |
ENST00000367871.1 ENST00000237264.4 |
TBPL1 |
TBP-like 1 |
| chr9_+_134269439 | 0.32 |
ENST00000405995.1 |
PRRC2B |
proline-rich coiled-coil 2B |
| chrX_+_23352133 | 0.31 |
ENST00000379361.4 |
PTCHD1 |
patched domain containing 1 |
| chr6_+_36410762 | 0.31 |
ENST00000483557.1 ENST00000498267.1 ENST00000544295.1 ENST00000449081.2 ENST00000536244.1 ENST00000460983.1 |
KCTD20 |
potassium channel tetramerization domain containing 20 |
| chr1_+_3370990 | 0.31 |
ENST00000378378.4 |
ARHGEF16 |
Rho guanine nucleotide exchange factor (GEF) 16 |
| chrX_-_131352152 | 0.31 |
ENST00000342983.2 |
RAP2C |
RAP2C, member of RAS oncogene family |
| chr2_+_65216462 | 0.31 |
ENST00000234256.3 |
SLC1A4 |
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr5_-_37835010 | 0.30 |
ENST00000510177.1 |
GDNF |
glial cell derived neurotrophic factor |
| chr11_-_60719213 | 0.30 |
ENST00000227880.3 |
SLC15A3 |
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr3_-_50540854 | 0.30 |
ENST00000423994.2 ENST00000424201.2 ENST00000479441.1 ENST00000429770.1 |
CACNA2D2 |
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
| chr3_+_184097836 | 0.30 |
ENST00000204604.1 ENST00000310236.3 |
CHRD |
chordin |
| chrX_+_41192595 | 0.30 |
ENST00000399959.2 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr17_-_78194147 | 0.29 |
ENST00000534910.1 ENST00000326317.6 |
SGSH |
N-sulfoglucosamine sulfohydrolase |
| chr11_-_64510409 | 0.29 |
ENST00000394429.1 ENST00000394428.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr22_+_19466980 | 0.29 |
ENST00000407835.1 ENST00000438587.1 |
CDC45 |
cell division cycle 45 |
| chr14_+_103243813 | 0.29 |
ENST00000560371.1 ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3 |
TNF receptor-associated factor 3 |
| chr19_+_5823813 | 0.29 |
ENST00000303212.2 |
NRTN |
neurturin |
| chr3_-_194991876 | 0.29 |
ENST00000310380.6 |
XXYLT1 |
xyloside xylosyltransferase 1 |
| chr11_+_64781575 | 0.28 |
ENST00000246747.4 ENST00000529384.1 |
ARL2 |
ADP-ribosylation factor-like 2 |
| chr11_+_64781657 | 0.28 |
ENST00000533729.1 |
ARL2 |
ADP-ribosylation factor-like 2 |
| chr22_-_26986045 | 0.28 |
ENST00000442495.1 ENST00000440953.1 ENST00000450022.1 ENST00000338754.4 |
TPST2 |
tyrosylprotein sulfotransferase 2 |
| chr3_+_184097905 | 0.28 |
ENST00000450923.1 |
CHRD |
chordin |
| chr2_+_10183651 | 0.27 |
ENST00000305883.1 |
KLF11 |
Kruppel-like factor 11 |
| chr17_-_41738931 | 0.27 |
ENST00000329168.3 ENST00000549132.1 |
MEOX1 |
mesenchyme homeobox 1 |
| chr10_+_88854926 | 0.27 |
ENST00000298784.1 ENST00000298786.4 |
FAM35A |
family with sequence similarity 35, member A |
| chr5_+_102455968 | 0.27 |
ENST00000358359.3 |
PPIP5K2 |
diphosphoinositol pentakisphosphate kinase 2 |
| chr8_+_90769967 | 0.26 |
ENST00000220751.4 |
RIPK2 |
receptor-interacting serine-threonine kinase 2 |
| chr7_+_128577972 | 0.26 |
ENST00000357234.5 ENST00000477535.1 ENST00000479582.1 ENST00000464557.1 ENST00000402030.2 |
IRF5 |
interferon regulatory factor 5 |
| chr6_+_143857949 | 0.26 |
ENST00000367584.4 |
PHACTR2 |
phosphatase and actin regulator 2 |
| chr12_-_51419924 | 0.25 |
ENST00000541174.2 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr16_+_29818857 | 0.25 |
ENST00000567444.1 |
MAZ |
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr17_-_4890919 | 0.25 |
ENST00000572543.1 ENST00000381311.5 ENST00000348066.3 ENST00000358183.4 |
CAMTA2 |
calmodulin binding transcription activator 2 |
| chr22_+_22735135 | 0.24 |
ENST00000390297.2 |
IGLV1-44 |
immunoglobulin lambda variable 1-44 |
| chr21_+_45285050 | 0.24 |
ENST00000291572.8 |
AGPAT3 |
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chrX_+_123095155 | 0.24 |
ENST00000371160.1 ENST00000435103.1 |
STAG2 |
stromal antigen 2 |
| chr19_-_50063907 | 0.24 |
ENST00000598296.1 |
NOSIP |
nitric oxide synthase interacting protein |
| chr4_-_149363662 | 0.24 |
ENST00000355292.3 ENST00000358102.3 |
NR3C2 |
nuclear receptor subfamily 3, group C, member 2 |
| chrX_+_24711997 | 0.23 |
ENST00000379068.3 ENST00000379059.3 |
POLA1 |
polymerase (DNA directed), alpha 1, catalytic subunit |
| chr6_+_89791507 | 0.23 |
ENST00000354922.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr9_+_100745615 | 0.23 |
ENST00000339399.4 |
ANP32B |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr9_-_88356789 | 0.23 |
ENST00000357081.3 ENST00000376081.4 ENST00000337006.4 ENST00000376109.3 |
AGTPBP1 |
ATP/GTP binding protein 1 |
| chr20_+_3869423 | 0.23 |
ENST00000497424.1 |
PANK2 |
pantothenate kinase 2 |
| chr9_+_100174232 | 0.23 |
ENST00000355295.4 |
TDRD7 |
tudor domain containing 7 |
| chr8_+_67976593 | 0.23 |
ENST00000262210.5 ENST00000412460.1 |
CSPP1 |
centrosome and spindle pole associated protein 1 |
| chrX_-_130423386 | 0.22 |
ENST00000370903.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr20_+_3870024 | 0.22 |
ENST00000610179.1 |
PANK2 |
pantothenate kinase 2 |
| chr2_+_134877740 | 0.22 |
ENST00000409645.1 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr14_-_64971288 | 0.22 |
ENST00000394715.1 |
ZBTB25 |
zinc finger and BTB domain containing 25 |
| chr9_-_34662651 | 0.22 |
ENST00000259631.4 |
CCL27 |
chemokine (C-C motif) ligand 27 |
| chr12_+_121163538 | 0.22 |
ENST00000242592.4 |
ACADS |
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr10_+_127408263 | 0.22 |
ENST00000337623.3 |
C10orf137 |
erythroid differentiation regulatory factor 1 |
| chr1_-_161147275 | 0.22 |
ENST00000319769.5 ENST00000367998.1 |
B4GALT3 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr16_+_2570340 | 0.22 |
ENST00000568263.1 ENST00000293971.6 ENST00000302956.4 ENST00000413459.3 ENST00000566706.1 ENST00000569879.1 |
AMDHD2 |
amidohydrolase domain containing 2 |
| chr10_-_105212141 | 0.21 |
ENST00000369788.3 |
CALHM2 |
calcium homeostasis modulator 2 |
| chr2_+_74781828 | 0.21 |
ENST00000340004.6 |
DOK1 |
docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
| chr7_-_140178726 | 0.21 |
ENST00000480552.1 |
MKRN1 |
makorin ring finger protein 1 |
| chrX_-_130423240 | 0.21 |
ENST00000370910.1 ENST00000370901.4 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr15_-_66790146 | 0.21 |
ENST00000316634.5 |
SNAPC5 |
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr3_+_38495333 | 0.21 |
ENST00000352511.4 |
ACVR2B |
activin A receptor, type IIB |
| chr12_-_48213568 | 0.21 |
ENST00000080059.7 ENST00000354334.3 ENST00000430670.1 ENST00000552960.1 ENST00000440293.1 |
HDAC7 |
histone deacetylase 7 |
| chr19_+_59055814 | 0.21 |
ENST00000594806.1 ENST00000253024.5 ENST00000341753.6 |
TRIM28 |
tripartite motif containing 28 |
| chr14_+_24641062 | 0.21 |
ENST00000311457.3 ENST00000557806.1 ENST00000559919.1 |
REC8 |
REC8 meiotic recombination protein |
| chr2_+_79740118 | 0.21 |
ENST00000496558.1 ENST00000451966.1 |
CTNNA2 |
catenin (cadherin-associated protein), alpha 2 |
| chr14_+_105886150 | 0.21 |
ENST00000331320.7 ENST00000406191.1 |
MTA1 |
metastasis associated 1 |
| chr12_-_51420108 | 0.21 |
ENST00000547198.1 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr20_-_62129163 | 0.20 |
ENST00000298049.7 |
EEF1A2 |
eukaryotic translation elongation factor 1 alpha 2 |
| chr4_+_115519577 | 0.20 |
ENST00000310836.6 |
UGT8 |
UDP glycosyltransferase 8 |
| chr3_-_9811595 | 0.20 |
ENST00000256460.3 |
CAMK1 |
calcium/calmodulin-dependent protein kinase I |
| chr2_-_11484710 | 0.20 |
ENST00000315872.6 |
ROCK2 |
Rho-associated, coiled-coil containing protein kinase 2 |
| chr10_+_14920843 | 0.20 |
ENST00000433779.1 ENST00000378325.3 ENST00000354919.6 ENST00000313519.5 ENST00000420416.1 |
SUV39H2 |
suppressor of variegation 3-9 homolog 2 (Drosophila) |
| chr4_+_2819883 | 0.20 |
ENST00000511747.1 ENST00000503393.2 |
SH3BP2 |
SH3-domain binding protein 2 |
| chr11_-_57004658 | 0.20 |
ENST00000606794.1 |
APLNR |
apelin receptor |
| chr12_-_48152853 | 0.19 |
ENST00000171000.4 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr16_+_58497567 | 0.19 |
ENST00000258187.5 |
NDRG4 |
NDRG family member 4 |
| chr14_-_95942173 | 0.19 |
ENST00000334258.5 ENST00000557275.1 ENST00000553340.1 |
SYNE3 |
spectrin repeat containing, nuclear envelope family member 3 |
| chr19_+_10362882 | 0.19 |
ENST00000393733.2 ENST00000588502.1 |
MRPL4 |
mitochondrial ribosomal protein L4 |
| chr1_+_3607228 | 0.19 |
ENST00000378285.1 ENST00000378280.1 ENST00000378288.4 |
TP73 |
tumor protein p73 |
| chr13_-_41345277 | 0.18 |
ENST00000323563.6 |
MRPS31 |
mitochondrial ribosomal protein S31 |
| chr20_-_62601218 | 0.18 |
ENST00000369888.1 |
ZNF512B |
zinc finger protein 512B |
| chr4_-_121993673 | 0.18 |
ENST00000379692.4 |
NDNF |
neuron-derived neurotrophic factor |
| chr1_+_28995231 | 0.18 |
ENST00000373816.1 |
GMEB1 |
glucocorticoid modulatory element binding protein 1 |
| chr3_+_38179969 | 0.18 |
ENST00000396334.3 ENST00000417037.2 ENST00000424893.1 ENST00000495303.1 ENST00000443433.2 ENST00000421516.1 |
MYD88 |
myeloid differentiation primary response 88 |
| chrX_+_51927919 | 0.18 |
ENST00000416960.1 |
MAGED4 |
melanoma antigen family D, 4 |
| chr6_+_44238203 | 0.18 |
ENST00000451188.2 |
TMEM151B |
transmembrane protein 151B |
| chr14_+_91580357 | 0.18 |
ENST00000298858.4 ENST00000521081.1 ENST00000520328.1 ENST00000256324.10 ENST00000524232.1 ENST00000522170.1 ENST00000519950.1 ENST00000523879.1 ENST00000521077.2 ENST00000518665.2 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chr14_+_91580708 | 0.18 |
ENST00000518868.1 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chr19_-_12992244 | 0.18 |
ENST00000538460.1 |
DNASE2 |
deoxyribonuclease II, lysosomal |
| chr6_+_15246501 | 0.18 |
ENST00000341776.2 |
JARID2 |
jumonji, AT rich interactive domain 2 |
| chr15_-_91565770 | 0.18 |
ENST00000535906.1 ENST00000333371.3 |
VPS33B |
vacuolar protein sorting 33 homolog B (yeast) |
| chr4_-_168155169 | 0.17 |
ENST00000534949.1 ENST00000535728.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr16_+_58059470 | 0.17 |
ENST00000219271.3 |
MMP15 |
matrix metallopeptidase 15 (membrane-inserted) |
| chr19_+_589893 | 0.17 |
ENST00000251287.2 |
HCN2 |
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr17_-_4871085 | 0.17 |
ENST00000575142.1 ENST00000206020.3 |
SPAG7 |
sperm associated antigen 7 |
| chr21_+_45725050 | 0.17 |
ENST00000403390.1 |
PFKL |
phosphofructokinase, liver |
| chr12_-_12503156 | 0.17 |
ENST00000543314.1 ENST00000396349.3 |
MANSC1 |
MANSC domain containing 1 |
| chr1_+_114471809 | 0.17 |
ENST00000426820.2 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr4_+_81951957 | 0.17 |
ENST00000282701.2 |
BMP3 |
bone morphogenetic protein 3 |
| chr10_-_105212059 | 0.17 |
ENST00000260743.5 |
CALHM2 |
calcium homeostasis modulator 2 |
| chr7_+_150020329 | 0.17 |
ENST00000323078.7 |
LRRC61 |
leucine rich repeat containing 61 |
| chr2_+_48757278 | 0.17 |
ENST00000404752.1 ENST00000406226.1 |
STON1 |
stonin 1 |
| chr12_-_121734489 | 0.17 |
ENST00000412367.2 ENST00000402834.4 ENST00000404169.3 |
CAMKK2 |
calcium/calmodulin-dependent protein kinase kinase 2, beta |
| chr1_+_146714291 | 0.17 |
ENST00000431239.1 ENST00000369259.3 ENST00000369258.4 ENST00000361293.5 |
CHD1L |
chromodomain helicase DNA binding protein 1-like |
| chr19_-_49314269 | 0.17 |
ENST00000545387.2 ENST00000316273.6 ENST00000402551.1 ENST00000598162.1 ENST00000599246.1 |
BCAT2 |
branched chain amino-acid transaminase 2, mitochondrial |
| chr7_+_73498118 | 0.17 |
ENST00000336180.2 |
LIMK1 |
LIM domain kinase 1 |
| chr16_+_29465822 | 0.16 |
ENST00000330181.5 ENST00000351581.4 |
SLX1B |
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr9_-_140351928 | 0.16 |
ENST00000339554.3 |
NSMF |
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr14_+_91580777 | 0.16 |
ENST00000525393.2 ENST00000428926.2 ENST00000517362.1 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chrX_-_153881842 | 0.16 |
ENST00000369585.3 ENST00000247306.4 |
CTAG2 |
cancer/testis antigen 2 |
| chrX_+_123094672 | 0.16 |
ENST00000354548.5 ENST00000458700.1 |
STAG2 |
stromal antigen 2 |
| chr11_+_47291193 | 0.16 |
ENST00000428807.1 ENST00000402799.1 ENST00000406482.1 ENST00000349238.3 ENST00000311027.5 ENST00000407859.3 ENST00000395344.3 ENST00000444117.1 |
MADD |
MAP-kinase activating death domain |
| chr1_+_28995258 | 0.16 |
ENST00000361872.4 ENST00000294409.2 |
GMEB1 |
glucocorticoid modulatory element binding protein 1 |
| chr2_-_64881018 | 0.16 |
ENST00000313349.3 |
SERTAD2 |
SERTA domain containing 2 |
| chr22_-_31328881 | 0.15 |
ENST00000445980.1 |
MORC2 |
MORC family CW-type zinc finger 2 |
| chr17_-_73178599 | 0.15 |
ENST00000578238.1 |
SUMO2 |
small ubiquitin-like modifier 2 |
| chr22_+_19705928 | 0.15 |
ENST00000383045.3 ENST00000438754.2 |
SEPT5 |
septin 5 |
| chr3_-_194090460 | 0.15 |
ENST00000428839.1 ENST00000347624.3 |
LRRC15 |
leucine rich repeat containing 15 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.2 | 0.7 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.2 | 0.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 2.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.4 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 5.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.9 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.7 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.5 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 2.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 1.0 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 1.0 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0031970 | organelle envelope lumen(GO:0031970) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 2.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 3.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.3 | 1.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.3 | 1.4 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.3 | 1.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.3 | 0.8 | GO:2000525 | dendritic cell dendrite assembly(GO:0097026) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.2 | 0.7 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.2 | 4.9 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 0.7 | GO:0031938 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 | 2.3 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 2.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.1 | 0.6 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.1 | 0.4 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.8 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.1 | 0.4 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.1 | 0.4 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.5 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.4 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.1 | 0.4 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.1 | 0.5 | GO:0021997 | response to chlorate(GO:0010157) neural plate axis specification(GO:0021997) |
| 0.1 | 1.1 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 | 0.3 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 1.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.1 | 0.7 | GO:0000101 | sulfur amino acid transport(GO:0000101) |
| 0.1 | 0.3 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.4 | GO:0048865 | stem cell fate commitment(GO:0048865) |
| 0.1 | 2.6 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.4 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.3 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.1 | 0.3 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) |
| 0.1 | 0.5 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.2 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.4 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.2 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 0.2 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 0.6 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 0.7 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.1 | 0.3 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.1 | 0.3 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.1 | 0.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.2 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.1 | GO:0002572 | pro-T cell differentiation(GO:0002572) |
| 0.1 | 0.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.4 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.1 | 0.2 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 | 0.4 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.5 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.9 | GO:0043306 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 1.3 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.5 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.2 | GO:0048617 | embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.4 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.2 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.0 | 0.3 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.3 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.4 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.2 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.1 | GO:0044571 | [2Fe-2S] cluster assembly(GO:0044571) |
| 0.0 | 0.1 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.0 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.0 | 0.1 | GO:0034471 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.5 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.2 | GO:0033490 | cholesterol biosynthetic process via desmosterol(GO:0033489) cholesterol biosynthetic process via lathosterol(GO:0033490) |
| 0.0 | 0.5 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.1 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.9 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.0 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 0.0 | 0.1 | GO:2000229 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.1 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.1 | GO:0021913 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) type B pancreatic cell maturation(GO:0072560) |
| 0.0 | 0.1 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0035802 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:1904431 | positive regulation of t-circle formation(GO:1904431) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.2 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.2 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.7 | GO:0002092 | positive regulation of receptor internalization(GO:0002092) |
| 0.0 | 0.7 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.5 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.5 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.6 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.4 | GO:0044342 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.7 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.7 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 0.1 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.1 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.2 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.2 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.0 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.0 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.0 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.2 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0060627 | regulation of vesicle-mediated transport(GO:0060627) |
| 0.0 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.1 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.2 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0015870 | acetylcholine transport(GO:0015870) |
| 0.0 | 0.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 1.0 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.1 | GO:0061047 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) regulation of branching involved in lung morphogenesis(GO:0061046) positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.0 | 3.1 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.0 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.0 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.0 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.0 | 0.1 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.0 | 0.2 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.4 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.4 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.1 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME P75NTR SIGNALS VIA NFKB | Genes involved in p75NTR signals via NF-kB |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 2.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 1.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.6 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.4 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.4 | 1.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.2 | 0.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 0.5 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.7 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.1 | 2.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.4 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.8 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.5 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 2.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.3 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.1 | 0.7 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.4 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.2 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 0.7 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.5 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.4 | GO:0015087 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.2 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 1.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.4 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.1 | 0.3 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.3 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.7 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.7 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.1 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.4 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.9 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 2.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0042578 | phosphoric ester hydrolase activity(GO:0042578) |
| 0.0 | 0.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.3 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.2 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 3.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0045309 | phosphotyrosine binding(GO:0001784) protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0005123 | death receptor binding(GO:0005123) |