Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
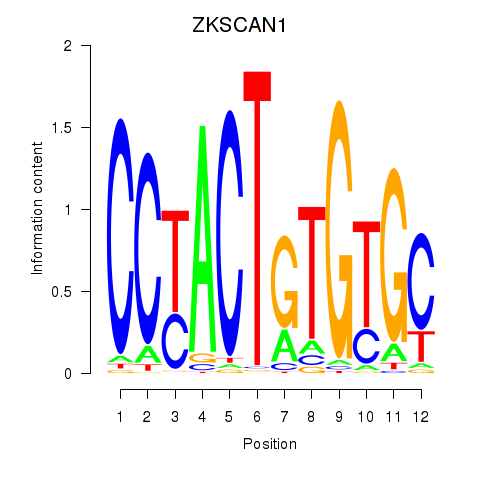
Results for ZKSCAN1
Z-value: 0.57
Transcription factors associated with ZKSCAN1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZKSCAN1
|
ENSG00000106261.12 | ZKSCAN1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZKSCAN1 | hg19_v2_chr7_+_99613195_99613206 | -0.63 | 8.5e-03 | Click! |
Activity profile of ZKSCAN1 motif
Sorted Z-values of ZKSCAN1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZKSCAN1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_4385230 | 2.35 |
ENST00000536537.1 |
CCND2 |
cyclin D2 |
| chr11_-_58343319 | 1.84 |
ENST00000395074.2 |
LPXN |
leupaxin |
| chr22_-_37640277 | 1.23 |
ENST00000401529.3 ENST00000249071.6 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr22_-_37640456 | 0.88 |
ENST00000405484.1 ENST00000441619.1 ENST00000406508.1 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr2_-_158345462 | 0.72 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr19_-_55660561 | 0.69 |
ENST00000587758.1 ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr19_+_39138320 | 0.62 |
ENST00000424234.2 ENST00000390009.3 ENST00000589528.1 |
ACTN4 |
actinin, alpha 4 |
| chr1_+_65613340 | 0.60 |
ENST00000546702.1 |
AK4 |
adenylate kinase 4 |
| chr20_-_56265680 | 0.58 |
ENST00000414037.1 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr5_-_134735568 | 0.48 |
ENST00000510038.1 ENST00000304332.4 |
H2AFY |
H2A histone family, member Y |
| chr8_-_131028869 | 0.48 |
ENST00000518283.1 ENST00000519110.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr1_+_46668994 | 0.47 |
ENST00000371980.3 |
LURAP1 |
leucine rich adaptor protein 1 |
| chrX_+_47441712 | 0.46 |
ENST00000218388.4 ENST00000377018.2 ENST00000456754.2 ENST00000377017.1 ENST00000441738.1 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
| chr9_-_112260531 | 0.42 |
ENST00000374541.2 ENST00000262539.3 |
PTPN3 |
protein tyrosine phosphatase, non-receptor type 3 |
| chr20_+_43343517 | 0.41 |
ENST00000372865.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr20_+_43343476 | 0.40 |
ENST00000372868.2 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr9_+_132934835 | 0.39 |
ENST00000372398.3 |
NCS1 |
neuronal calcium sensor 1 |
| chr5_-_127873659 | 0.37 |
ENST00000262464.4 |
FBN2 |
fibrillin 2 |
| chr1_+_65613217 | 0.36 |
ENST00000545314.1 |
AK4 |
adenylate kinase 4 |
| chr22_+_29702572 | 0.30 |
ENST00000407647.2 ENST00000416823.1 ENST00000428622.1 |
GAS2L1 |
growth arrest-specific 2 like 1 |
| chr15_-_34635314 | 0.27 |
ENST00000557912.1 ENST00000328848.4 |
NOP10 |
NOP10 ribonucleoprotein |
| chr11_+_70049269 | 0.26 |
ENST00000301838.4 |
FADD |
Fas (TNFRSF6)-associated via death domain |
| chr19_+_39138271 | 0.24 |
ENST00000252699.2 |
ACTN4 |
actinin, alpha 4 |
| chr19_-_3062465 | 0.24 |
ENST00000327141.4 |
AES |
amino-terminal enhancer of split |
| chr5_+_174151536 | 0.23 |
ENST00000239243.6 ENST00000507785.1 |
MSX2 |
msh homeobox 2 |
| chr22_+_21921994 | 0.23 |
ENST00000545681.1 |
UBE2L3 |
ubiquitin-conjugating enzyme E2L 3 |
| chr14_+_103851712 | 0.22 |
ENST00000440884.3 ENST00000416682.2 ENST00000429436.2 ENST00000303622.9 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
| chr11_-_118134997 | 0.22 |
ENST00000278937.2 |
MPZL2 |
myelin protein zero-like 2 |
| chr1_-_160549235 | 0.21 |
ENST00000368054.3 ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84 |
CD84 molecule |
| chr10_+_112679301 | 0.20 |
ENST00000265277.5 ENST00000369452.4 |
SHOC2 |
soc-2 suppressor of clear homolog (C. elegans) |
| chr5_+_135394840 | 0.20 |
ENST00000503087.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr16_+_19125252 | 0.20 |
ENST00000566735.1 ENST00000381440.3 |
ITPRIPL2 |
inositol 1,4,5-trisphosphate receptor interacting protein-like 2 |
| chr19_-_3062881 | 0.20 |
ENST00000586742.1 |
AES |
amino-terminal enhancer of split |
| chr15_-_49103235 | 0.20 |
ENST00000380950.2 |
CEP152 |
centrosomal protein 152kDa |
| chr2_+_74120094 | 0.19 |
ENST00000409731.3 ENST00000345517.3 ENST00000409918.1 ENST00000442912.1 ENST00000409624.1 |
ACTG2 |
actin, gamma 2, smooth muscle, enteric |
| chr17_-_73150599 | 0.19 |
ENST00000392566.2 ENST00000581874.1 |
HN1 |
hematological and neurological expressed 1 |
| chr17_+_26800756 | 0.19 |
ENST00000537681.1 |
SLC13A2 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr20_+_44098385 | 0.18 |
ENST00000217425.5 ENST00000339946.3 |
WFDC2 |
WAP four-disulfide core domain 2 |
| chr20_+_44098346 | 0.18 |
ENST00000372676.3 |
WFDC2 |
WAP four-disulfide core domain 2 |
| chrX_+_109245863 | 0.18 |
ENST00000372072.3 |
TMEM164 |
transmembrane protein 164 |
| chr22_+_33197683 | 0.18 |
ENST00000266085.6 |
TIMP3 |
TIMP metallopeptidase inhibitor 3 |
| chr2_-_183387430 | 0.18 |
ENST00000410103.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr2_-_37193606 | 0.18 |
ENST00000379213.2 ENST00000263918.4 |
STRN |
striatin, calmodulin binding protein |
| chr17_+_26800648 | 0.17 |
ENST00000545060.1 |
SLC13A2 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr19_+_10196781 | 0.17 |
ENST00000253110.11 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr17_-_26220366 | 0.17 |
ENST00000460380.2 ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9 RP1-66C13.4 |
LYR motif containing 9 Uncharacterized protein |
| chr19_-_3063099 | 0.16 |
ENST00000221561.8 |
AES |
amino-terminal enhancer of split |
| chr1_-_36615051 | 0.16 |
ENST00000373163.1 |
TRAPPC3 |
trafficking protein particle complex 3 |
| chr6_+_35265586 | 0.16 |
ENST00000542066.1 ENST00000316637.5 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
| chr17_-_73150629 | 0.15 |
ENST00000356033.4 ENST00000405458.3 ENST00000409753.3 |
HN1 |
hematological and neurological expressed 1 |
| chr1_-_36615065 | 0.14 |
ENST00000373166.3 ENST00000373159.1 ENST00000373162.1 |
TRAPPC3 |
trafficking protein particle complex 3 |
| chr19_+_13135790 | 0.13 |
ENST00000358552.3 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr12_+_122326662 | 0.13 |
ENST00000261817.2 ENST00000538613.1 ENST00000542602.1 |
PSMD9 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr5_-_134734901 | 0.13 |
ENST00000312469.4 ENST00000423969.2 |
H2AFY |
H2A histone family, member Y |
| chr15_-_80215984 | 0.12 |
ENST00000485386.1 ENST00000479961.1 |
ST20 ST20-MTHFS |
suppressor of tumorigenicity 20 ST20-MTHFS readthrough |
| chr17_+_27071002 | 0.12 |
ENST00000262395.5 ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4 |
TNF receptor-associated factor 4 |
| chr19_-_19051993 | 0.12 |
ENST00000594794.1 ENST00000355887.6 ENST00000392351.3 ENST00000596482.1 |
HOMER3 |
homer homolog 3 (Drosophila) |
| chr7_+_87563557 | 0.11 |
ENST00000439864.1 ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22 |
ADAM metallopeptidase domain 22 |
| chr16_+_2076869 | 0.11 |
ENST00000424542.2 ENST00000432365.2 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr6_+_151815143 | 0.11 |
ENST00000239374.7 ENST00000367290.5 |
CCDC170 |
coiled-coil domain containing 170 |
| chr10_+_102222798 | 0.11 |
ENST00000343737.5 |
WNT8B |
wingless-type MMTV integration site family, member 8B |
| chr11_-_73720276 | 0.10 |
ENST00000348534.4 |
UCP3 |
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr12_+_122326630 | 0.10 |
ENST00000541212.1 ENST00000340175.5 |
PSMD9 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr4_-_176733897 | 0.10 |
ENST00000393658.2 |
GPM6A |
glycoprotein M6A |
| chr3_+_53528659 | 0.10 |
ENST00000350061.5 |
CACNA1D |
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr2_+_113931513 | 0.10 |
ENST00000245796.6 ENST00000441564.3 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
| chr16_+_3550924 | 0.09 |
ENST00000576634.1 ENST00000574369.1 ENST00000341633.5 ENST00000417763.2 ENST00000571025.1 |
CLUAP1 |
clusterin associated protein 1 |
| chr19_+_18794470 | 0.09 |
ENST00000321949.8 ENST00000338797.6 |
CRTC1 |
CREB regulated transcription coactivator 1 |
| chr17_+_75372165 | 0.08 |
ENST00000427674.2 |
SEPT9 |
septin 9 |
| chrX_-_15402498 | 0.08 |
ENST00000297904.3 |
FIGF |
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr10_-_88281494 | 0.08 |
ENST00000298767.5 |
WAPAL |
wings apart-like homolog (Drosophila) |
| chr11_-_73720122 | 0.08 |
ENST00000426995.2 |
UCP3 |
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr19_+_13135731 | 0.07 |
ENST00000587260.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr1_-_155224751 | 0.07 |
ENST00000350210.2 ENST00000368368.3 |
FAM189B |
family with sequence similarity 189, member B |
| chr1_+_32083301 | 0.06 |
ENST00000403528.2 |
HCRTR1 |
hypocretin (orexin) receptor 1 |
| chr2_+_101179152 | 0.06 |
ENST00000264254.6 |
PDCL3 |
phosducin-like 3 |
| chr7_+_87563458 | 0.06 |
ENST00000398204.4 |
ADAM22 |
ADAM metallopeptidase domain 22 |
| chr10_-_112678976 | 0.06 |
ENST00000448814.1 |
BBIP1 |
BBSome interacting protein 1 |
| chr15_+_75494214 | 0.06 |
ENST00000394987.4 |
C15orf39 |
chromosome 15 open reading frame 39 |
| chr15_-_49103184 | 0.05 |
ENST00000399334.3 ENST00000325747.5 |
CEP152 |
centrosomal protein 152kDa |
| chr19_-_15918936 | 0.05 |
ENST00000334920.2 |
OR10H1 |
olfactory receptor, family 10, subfamily H, member 1 |
| chr2_+_1488435 | 0.04 |
ENST00000446278.1 ENST00000469607.1 |
TPO |
thyroid peroxidase |
| chr17_+_18163848 | 0.04 |
ENST00000323019.4 ENST00000578174.1 ENST00000395704.4 ENST00000395703.4 ENST00000578621.1 ENST00000579341.1 |
MIEF2 |
mitochondrial elongation factor 2 |
| chr1_-_177134024 | 0.04 |
ENST00000367654.3 |
ASTN1 |
astrotactin 1 |
| chr10_-_126432619 | 0.04 |
ENST00000337318.3 |
FAM53B |
family with sequence similarity 53, member B |
| chr2_+_27070964 | 0.04 |
ENST00000288699.6 |
DPYSL5 |
dihydropyrimidinase-like 5 |
| chr18_+_52495426 | 0.03 |
ENST00000262094.5 |
RAB27B |
RAB27B, member RAS oncogene family |
| chr19_+_3178736 | 0.03 |
ENST00000246115.3 |
S1PR4 |
sphingosine-1-phosphate receptor 4 |
| chr19_+_51630287 | 0.03 |
ENST00000599948.1 |
SIGLEC9 |
sialic acid binding Ig-like lectin 9 |
| chr6_+_32944119 | 0.03 |
ENST00000606059.1 |
BRD2 |
bromodomain containing 2 |
| chr11_+_72975578 | 0.03 |
ENST00000393592.2 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr10_-_112678904 | 0.03 |
ENST00000423273.1 ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1 |
BBSome interacting protein 1 |
| chr11_+_72975524 | 0.03 |
ENST00000540342.1 ENST00000542092.1 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr20_-_3644046 | 0.02 |
ENST00000290417.2 ENST00000319242.3 |
GFRA4 |
GDNF family receptor alpha 4 |
| chr4_+_39460689 | 0.02 |
ENST00000381846.1 |
LIAS |
lipoic acid synthetase |
| chr3_+_184081213 | 0.02 |
ENST00000429568.1 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr11_+_72975559 | 0.02 |
ENST00000349767.2 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr6_-_30640811 | 0.01 |
ENST00000376442.3 |
DHX16 |
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr3_+_184081175 | 0.01 |
ENST00000452961.1 ENST00000296223.3 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr3_-_195310802 | 0.01 |
ENST00000421243.1 ENST00000453131.1 |
APOD |
apolipoprotein D |
| chr1_-_51796987 | 0.01 |
ENST00000262676.5 |
TTC39A |
tetratricopeptide repeat domain 39A |
| chr1_+_209859510 | 0.01 |
ENST00000367028.2 ENST00000261465.1 |
HSD11B1 |
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr19_+_850985 | 0.00 |
ENST00000590230.1 |
ELANE |
elastase, neutrophil expressed |
| chr3_+_63898275 | 0.00 |
ENST00000538065.1 |
ATXN7 |
ataxin 7 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.2 | 1.8 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.2 | 0.6 | GO:1901837 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.2 | 2.1 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.2 | 1.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 2.4 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 1.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.5 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.2 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.1 | 0.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.3 | GO:0072683 | T cell extravasation(GO:0072683) |
| 0.1 | 0.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.3 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.2 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.6 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.3 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.4 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.1 | 0.4 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.1 | 0.5 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.6 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.3 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.6 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 0.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.3 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 1.8 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 2.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.3 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 2.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |