Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
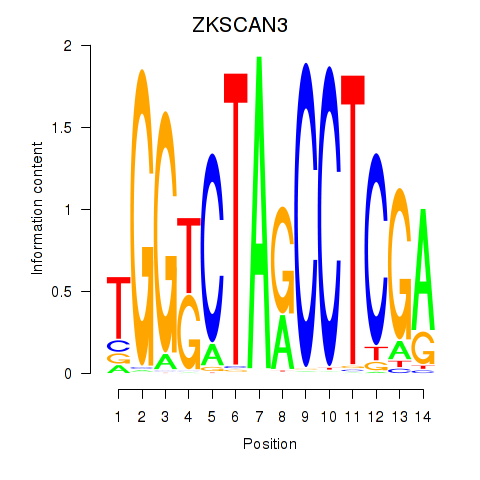
Results for ZKSCAN3
Z-value: 0.44
Transcription factors associated with ZKSCAN3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZKSCAN3
|
ENSG00000189298.9 | ZKSCAN3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZKSCAN3 | hg19_v2_chr6_+_28317685_28317761 | 0.03 | 9.1e-01 | Click! |
Activity profile of ZKSCAN3 motif
Sorted Z-values of ZKSCAN3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZKSCAN3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_41624685 | 0.69 |
ENST00000560640.1 ENST00000220514.3 |
OIP5 |
Opa interacting protein 5 |
| chr15_+_41624892 | 0.65 |
ENST00000260359.6 ENST00000450318.1 ENST00000450592.2 ENST00000559596.1 ENST00000414849.2 ENST00000560747.1 ENST00000560177.1 |
NUSAP1 |
nucleolar and spindle associated protein 1 |
| chr12_+_100867486 | 0.51 |
ENST00000548884.1 |
NR1H4 |
nuclear receptor subfamily 1, group H, member 4 |
| chr12_+_100867694 | 0.46 |
ENST00000392986.3 ENST00000549996.1 |
NR1H4 |
nuclear receptor subfamily 1, group H, member 4 |
| chrX_+_23352133 | 0.43 |
ENST00000379361.4 |
PTCHD1 |
patched domain containing 1 |
| chr12_+_109535923 | 0.43 |
ENST00000336865.2 |
UNG |
uracil-DNA glycosylase |
| chr12_+_109535373 | 0.40 |
ENST00000242576.2 |
UNG |
uracil-DNA glycosylase |
| chr1_+_66258846 | 0.37 |
ENST00000341517.4 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
| chr14_-_106926724 | 0.35 |
ENST00000434710.1 |
IGHV3-43 |
immunoglobulin heavy variable 3-43 |
| chr8_+_123793633 | 0.23 |
ENST00000314393.4 |
ZHX2 |
zinc fingers and homeoboxes 2 |
| chr5_+_65018017 | 0.23 |
ENST00000380985.5 ENST00000502464.1 |
NLN |
neurolysin (metallopeptidase M3 family) |
| chr1_+_226250379 | 0.23 |
ENST00000366815.3 ENST00000366814.3 |
H3F3A |
H3 histone, family 3A |
| chr1_+_93913665 | 0.22 |
ENST00000271234.7 ENST00000370256.4 ENST00000260506.8 |
FNBP1L |
formin binding protein 1-like |
| chr19_-_49258606 | 0.20 |
ENST00000310160.3 |
FUT1 |
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) |
| chr1_+_156123359 | 0.18 |
ENST00000368284.1 ENST00000368286.2 ENST00000438830.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr17_-_60142609 | 0.18 |
ENST00000397786.2 |
MED13 |
mediator complex subunit 13 |
| chr1_+_156123318 | 0.18 |
ENST00000368285.3 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr13_-_111567353 | 0.17 |
ENST00000310847.4 ENST00000267339.2 ENST00000375758.5 |
ANKRD10 |
ankyrin repeat domain 10 |
| chr19_+_54372639 | 0.17 |
ENST00000391769.2 |
MYADM |
myeloid-associated differentiation marker |
| chr19_+_49258775 | 0.17 |
ENST00000593756.1 |
FGF21 |
fibroblast growth factor 21 |
| chr10_+_21823079 | 0.16 |
ENST00000377100.3 ENST00000377072.3 ENST00000446906.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr1_+_93913713 | 0.15 |
ENST00000604705.1 ENST00000370253.2 |
FNBP1L |
formin binding protein 1-like |
| chr6_+_32936353 | 0.15 |
ENST00000374825.4 |
BRD2 |
bromodomain containing 2 |
| chr15_+_74610894 | 0.14 |
ENST00000558821.1 ENST00000268082.4 |
CCDC33 |
coiled-coil domain containing 33 |
| chr16_+_2479390 | 0.13 |
ENST00000397066.4 |
CCNF |
cyclin F |
| chr18_-_74728998 | 0.12 |
ENST00000359645.3 ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP |
myelin basic protein |
| chr10_+_21823243 | 0.12 |
ENST00000307729.7 ENST00000377091.2 |
MLLT10 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr6_-_35656712 | 0.11 |
ENST00000357266.4 ENST00000542713.1 |
FKBP5 |
FK506 binding protein 5 |
| chr12_-_3982511 | 0.09 |
ENST00000427057.2 ENST00000228820.4 |
PARP11 |
poly (ADP-ribose) polymerase family, member 11 |
| chr5_-_65017921 | 0.09 |
ENST00000381007.4 |
SGTB |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr2_-_175351744 | 0.09 |
ENST00000295500.4 ENST00000392552.2 ENST00000392551.2 |
GPR155 |
G protein-coupled receptor 155 |
| chr11_-_64013288 | 0.07 |
ENST00000542235.1 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr6_-_31508304 | 0.06 |
ENST00000376177.2 |
DDX39B |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr1_+_3598871 | 0.06 |
ENST00000603362.1 ENST00000604479.1 |
TP73 |
tumor protein p73 |
| chr1_-_115259337 | 0.05 |
ENST00000369535.4 |
NRAS |
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr12_-_3982548 | 0.05 |
ENST00000397096.2 ENST00000447133.3 ENST00000450737.2 |
PARP11 |
poly (ADP-ribose) polymerase family, member 11 |
| chr22_-_50312052 | 0.04 |
ENST00000330817.6 |
ALG12 |
ALG12, alpha-1,6-mannosyltransferase |
| chr11_-_67272794 | 0.03 |
ENST00000436757.2 ENST00000356404.3 |
PITPNM1 |
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr14_-_106069247 | 0.03 |
ENST00000479229.1 |
RP11-731F5.1 |
RP11-731F5.1 |
| chr2_+_191745560 | 0.03 |
ENST00000338435.4 |
GLS |
glutaminase |
| chr11_+_66610883 | 0.03 |
ENST00000309657.3 ENST00000524506.1 |
RCE1 |
Ras converting CAAX endopeptidase 1 |
| chr1_-_19811132 | 0.02 |
ENST00000433834.1 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
| chrX_+_153775821 | 0.01 |
ENST00000263518.6 ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr1_-_28415075 | 0.01 |
ENST00000373863.3 ENST00000436342.2 ENST00000540618.1 ENST00000545175.1 |
EYA3 |
eyes absent homolog 3 (Drosophila) |
| chr12_-_113573495 | 0.00 |
ENST00000446861.3 |
RASAL1 |
RAS protein activator like 1 (GAP1 like) |
| chr22_+_50312316 | 0.00 |
ENST00000328268.4 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.1 | 0.8 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:2000213 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.1 | 0.2 | GO:1902809 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.1 | 0.8 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.1 | 0.2 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.7 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.4 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.7 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.2 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.4 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 1.0 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |