Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
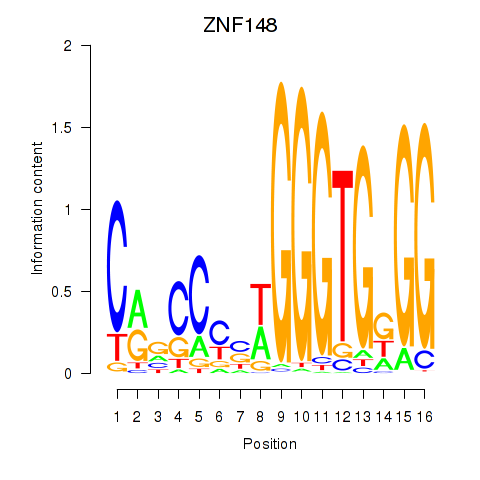
Results for ZNF148
Z-value: 1.10
Transcription factors associated with ZNF148
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF148
|
ENSG00000163848.14 | ZNF148 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF148 | hg19_v2_chr3_-_125094093_125094198 | -0.19 | 4.8e-01 | Click! |
Activity profile of ZNF148 motif
Sorted Z-values of ZNF148 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF148
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_57976544 | 3.24 |
ENST00000295666.4 ENST00000537922.1 |
IGFBP7 |
insulin-like growth factor binding protein 7 |
| chr2_+_173292280 | 2.94 |
ENST00000264107.7 |
ITGA6 |
integrin, alpha 6 |
| chr2_-_161349909 | 2.85 |
ENST00000392753.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr2_+_173292390 | 2.77 |
ENST00000442250.1 ENST00000458358.1 ENST00000409080.1 |
ITGA6 |
integrin, alpha 6 |
| chr2_+_173292301 | 2.70 |
ENST00000264106.6 ENST00000375221.2 ENST00000343713.4 |
ITGA6 |
integrin, alpha 6 |
| chr19_-_18717627 | 2.66 |
ENST00000392386.3 |
CRLF1 |
cytokine receptor-like factor 1 |
| chr19_+_41725088 | 2.40 |
ENST00000301178.4 |
AXL |
AXL receptor tyrosine kinase |
| chr11_-_2160180 | 2.39 |
ENST00000381406.4 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr11_-_2160611 | 2.30 |
ENST00000416167.2 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr5_+_52776449 | 2.17 |
ENST00000396947.3 |
FST |
follistatin |
| chr5_+_52776228 | 2.17 |
ENST00000256759.3 |
FST |
follistatin |
| chr2_-_161350305 | 2.04 |
ENST00000348849.3 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
| chr20_-_56285595 | 1.87 |
ENST00000395816.3 ENST00000347215.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr7_+_100770328 | 1.86 |
ENST00000223095.4 ENST00000445463.2 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr7_-_80548667 | 1.80 |
ENST00000265361.3 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr5_-_172756506 | 1.75 |
ENST00000265087.4 |
STC2 |
stanniocalcin 2 |
| chr10_+_48255253 | 1.73 |
ENST00000357718.4 ENST00000344416.5 ENST00000456111.2 ENST00000374258.3 |
ANXA8 AL591684.1 |
annexin A8 Protein LOC100996760 |
| chr10_-_47173994 | 1.67 |
ENST00000414655.2 ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1 LINC00842 |
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr12_+_6309517 | 1.65 |
ENST00000382519.4 ENST00000009180.4 |
CD9 |
CD9 molecule |
| chr10_+_47746929 | 1.63 |
ENST00000340243.6 ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2 AL603965.1 |
annexin A8-like 2 Protein LOC100996760 |
| chr19_-_50143452 | 1.56 |
ENST00000246792.3 |
RRAS |
related RAS viral (r-ras) oncogene homolog |
| chr3_+_57994127 | 1.52 |
ENST00000490882.1 ENST00000295956.4 ENST00000358537.3 ENST00000429972.2 ENST00000348383.5 ENST00000357272.4 |
FLNB |
filamin B, beta |
| chr4_+_75311019 | 1.51 |
ENST00000502307.1 |
AREG |
amphiregulin |
| chr4_+_75480629 | 1.44 |
ENST00000380846.3 |
AREGB |
amphiregulin B |
| chrX_+_114795489 | 1.44 |
ENST00000355899.3 ENST00000537301.1 ENST00000289290.3 |
PLS3 |
plastin 3 |
| chr3_+_159557637 | 1.43 |
ENST00000445224.2 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr19_-_19049791 | 1.41 |
ENST00000594439.1 ENST00000221222.11 |
HOMER3 |
homer homolog 3 (Drosophila) |
| chr16_+_8806800 | 1.38 |
ENST00000561870.1 ENST00000396600.2 |
ABAT |
4-aminobutyrate aminotransferase |
| chr19_-_51522955 | 1.38 |
ENST00000358789.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr11_+_131781290 | 1.37 |
ENST00000425719.2 ENST00000374784.1 |
NTM |
neurotrimin |
| chr13_+_98794810 | 1.32 |
ENST00000595437.1 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr7_+_17338239 | 1.26 |
ENST00000242057.4 |
AHR |
aryl hydrocarbon receptor |
| chr22_+_38093005 | 1.23 |
ENST00000406386.3 |
TRIOBP |
TRIO and F-actin binding protein |
| chr4_+_75310851 | 1.23 |
ENST00000395748.3 ENST00000264487.2 |
AREG |
amphiregulin |
| chr14_+_105331596 | 1.23 |
ENST00000556508.1 ENST00000414716.3 ENST00000453495.1 ENST00000418279.1 |
CEP170B |
centrosomal protein 170B |
| chrX_+_17755563 | 1.22 |
ENST00000380045.3 ENST00000380041.3 ENST00000380043.3 ENST00000398080.1 |
SCML1 |
sex comb on midleg-like 1 (Drosophila) |
| chr4_+_6271558 | 1.17 |
ENST00000503569.1 ENST00000226760.1 |
WFS1 |
Wolfram syndrome 1 (wolframin) |
| chr19_+_38755042 | 1.14 |
ENST00000301244.7 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
| chr9_+_116327326 | 1.10 |
ENST00000342620.5 |
RGS3 |
regulator of G-protein signaling 3 |
| chr11_+_12696102 | 1.04 |
ENST00000527636.1 ENST00000527376.1 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr12_-_52967600 | 1.01 |
ENST00000549343.1 ENST00000305620.2 |
KRT74 |
keratin 74 |
| chr9_+_131314859 | 1.00 |
ENST00000358161.5 ENST00000372731.4 ENST00000372739.3 |
SPTAN1 |
spectrin, alpha, non-erythrocytic 1 |
| chr17_+_2699697 | 0.99 |
ENST00000254695.8 ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
| chr2_+_201171372 | 0.99 |
ENST00000409140.3 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr10_-_118764862 | 0.98 |
ENST00000260777.10 |
KIAA1598 |
KIAA1598 |
| chr5_+_76114758 | 0.97 |
ENST00000514165.1 ENST00000296677.4 |
F2RL1 |
coagulation factor II (thrombin) receptor-like 1 |
| chr1_-_201346761 | 0.97 |
ENST00000455702.1 ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2 |
troponin T type 2 (cardiac) |
| chr10_+_134000404 | 0.95 |
ENST00000338492.4 ENST00000368629.1 |
DPYSL4 |
dihydropyrimidinase-like 4 |
| chrX_+_73641286 | 0.92 |
ENST00000587091.1 |
SLC16A2 |
solute carrier family 16, member 2 (thyroid hormone transporter) |
| chr8_+_32406179 | 0.92 |
ENST00000405005.3 |
NRG1 |
neuregulin 1 |
| chr17_-_64188177 | 0.92 |
ENST00000535342.2 |
CEP112 |
centrosomal protein 112kDa |
| chr2_-_20424844 | 0.91 |
ENST00000403076.1 ENST00000254351.4 |
SDC1 |
syndecan 1 |
| chrX_+_152990302 | 0.90 |
ENST00000218104.3 |
ABCD1 |
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr1_-_11714700 | 0.90 |
ENST00000354287.4 |
FBXO2 |
F-box protein 2 |
| chr16_+_6069586 | 0.88 |
ENST00000547372.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr3_+_50192499 | 0.87 |
ENST00000413852.1 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr17_-_1394940 | 0.87 |
ENST00000570984.2 ENST00000361007.2 |
MYO1C |
myosin IC |
| chr12_-_124457371 | 0.86 |
ENST00000238156.3 ENST00000545037.1 |
CCDC92 |
coiled-coil domain containing 92 |
| chrX_+_48367338 | 0.86 |
ENST00000359882.4 ENST00000537758.1 ENST00000367574.4 ENST00000355961.4 ENST00000489940.1 ENST00000361988.3 |
PORCN |
porcupine homolog (Drosophila) |
| chr14_+_105941118 | 0.85 |
ENST00000550577.1 ENST00000538259.2 |
CRIP2 |
cysteine-rich protein 2 |
| chr16_-_70719925 | 0.85 |
ENST00000338779.6 |
MTSS1L |
metastasis suppressor 1-like |
| chr3_-_52001448 | 0.85 |
ENST00000461554.1 ENST00000395013.3 ENST00000428823.2 ENST00000483411.1 ENST00000461544.1 ENST00000355852.2 |
PCBP4 |
poly(rC) binding protein 4 |
| chr13_+_102104980 | 0.84 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr5_+_125758813 | 0.83 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr1_-_95007193 | 0.82 |
ENST00000370207.4 ENST00000334047.7 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
| chr17_-_39968855 | 0.82 |
ENST00000355468.3 ENST00000590496.1 |
LEPREL4 |
leprecan-like 4 |
| chr3_+_50192537 | 0.81 |
ENST00000002829.3 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr16_-_4987065 | 0.77 |
ENST00000590782.2 ENST00000345988.2 |
PPL |
periplakin |
| chr21_-_28338732 | 0.76 |
ENST00000284987.5 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
| chr17_+_7348374 | 0.75 |
ENST00000306071.2 ENST00000572857.1 |
CHRNB1 |
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr13_+_102104952 | 0.73 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr12_-_90102594 | 0.73 |
ENST00000428670.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr8_-_67525473 | 0.73 |
ENST00000522677.3 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr10_-_99447024 | 0.71 |
ENST00000370626.3 |
AVPI1 |
arginine vasopressin-induced 1 |
| chr1_-_43232649 | 0.65 |
ENST00000372526.2 ENST00000236040.4 ENST00000296388.5 ENST00000397054.3 |
LEPRE1 |
leucine proline-enriched proteoglycan (leprecan) 1 |
| chr4_+_3768075 | 0.63 |
ENST00000509482.1 ENST00000330055.5 |
ADRA2C |
adrenoceptor alpha 2C |
| chr10_+_104503727 | 0.63 |
ENST00000448841.1 |
WBP1L |
WW domain binding protein 1-like |
| chr2_+_64681219 | 0.61 |
ENST00000238875.5 |
LGALSL |
lectin, galactoside-binding-like |
| chr22_-_38669030 | 0.61 |
ENST00000361906.3 |
TMEM184B |
transmembrane protein 184B |
| chrX_+_107069063 | 0.61 |
ENST00000262843.6 |
MID2 |
midline 2 |
| chr2_+_220143989 | 0.59 |
ENST00000336576.5 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr19_-_51523275 | 0.58 |
ENST00000309958.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr5_-_176900610 | 0.57 |
ENST00000477391.2 ENST00000393565.1 ENST00000309007.5 |
DBN1 |
drebrin 1 |
| chr1_+_183155373 | 0.57 |
ENST00000493293.1 ENST00000264144.4 |
LAMC2 |
laminin, gamma 2 |
| chr3_-_48130314 | 0.57 |
ENST00000439356.1 ENST00000395734.3 ENST00000426837.2 |
MAP4 |
microtubule-associated protein 4 |
| chr16_+_89238149 | 0.56 |
ENST00000289746.2 |
CDH15 |
cadherin 15, type 1, M-cadherin (myotubule) |
| chr17_+_7348658 | 0.55 |
ENST00000570557.1 ENST00000536404.2 ENST00000576360.1 |
CHRNB1 |
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr19_-_51523412 | 0.54 |
ENST00000391805.1 ENST00000599077.1 |
KLK10 |
kallikrein-related peptidase 10 |
| chr11_+_12132117 | 0.53 |
ENST00000256194.4 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr17_-_62658186 | 0.52 |
ENST00000262435.9 |
SMURF2 |
SMAD specific E3 ubiquitin protein ligase 2 |
| chr11_+_1944054 | 0.52 |
ENST00000397301.1 ENST00000397304.2 ENST00000446240.1 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
| chr1_+_145576007 | 0.51 |
ENST00000369298.1 |
PIAS3 |
protein inhibitor of activated STAT, 3 |
| chr8_-_25315905 | 0.51 |
ENST00000221200.4 |
KCTD9 |
potassium channel tetramerization domain containing 9 |
| chr7_+_94537542 | 0.50 |
ENST00000433881.1 |
PPP1R9A |
protein phosphatase 1, regulatory subunit 9A |
| chr5_+_71403061 | 0.50 |
ENST00000512974.1 ENST00000296755.7 |
MAP1B |
microtubule-associated protein 1B |
| chr10_+_73724123 | 0.49 |
ENST00000373115.4 |
CHST3 |
carbohydrate (chondroitin 6) sulfotransferase 3 |
| chr16_+_6069664 | 0.49 |
ENST00000422070.4 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr8_-_145018080 | 0.49 |
ENST00000354589.3 |
PLEC |
plectin |
| chr2_+_220144052 | 0.48 |
ENST00000425450.1 ENST00000392086.4 ENST00000421532.1 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr6_+_30130969 | 0.47 |
ENST00000376694.4 |
TRIM15 |
tripartite motif containing 15 |
| chr2_-_220408430 | 0.47 |
ENST00000243776.6 |
CHPF |
chondroitin polymerizing factor |
| chr1_+_145575980 | 0.47 |
ENST00000393045.2 |
PIAS3 |
protein inhibitor of activated STAT, 3 |
| chr2_+_128848740 | 0.47 |
ENST00000375990.3 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr9_+_19049372 | 0.46 |
ENST00000380527.1 |
RRAGA |
Ras-related GTP binding A |
| chr17_+_46126135 | 0.46 |
ENST00000361665.3 ENST00000585062.1 |
NFE2L1 |
nuclear factor, erythroid 2-like 1 |
| chr9_+_4679555 | 0.45 |
ENST00000381858.1 ENST00000381854.3 |
CDC37L1 |
cell division cycle 37-like 1 |
| chr2_+_64681103 | 0.45 |
ENST00000464281.1 |
LGALSL |
lectin, galactoside-binding-like |
| chr2_+_128848881 | 0.44 |
ENST00000259253.6 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr17_-_79818354 | 0.44 |
ENST00000576541.1 ENST00000576380.1 ENST00000571617.1 ENST00000576052.1 ENST00000576390.1 ENST00000573778.2 ENST00000439918.2 ENST00000574914.1 ENST00000331483.4 |
P4HB |
prolyl 4-hydroxylase, beta polypeptide |
| chr19_-_36297632 | 0.44 |
ENST00000588266.2 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chr13_-_96296944 | 0.43 |
ENST00000361396.2 ENST00000376829.2 |
DZIP1 |
DAZ interacting zinc finger protein 1 |
| chrX_+_47078069 | 0.43 |
ENST00000357227.4 ENST00000519758.1 ENST00000520893.1 ENST00000517426.1 |
CDK16 |
cyclin-dependent kinase 16 |
| chr6_+_89790459 | 0.43 |
ENST00000369472.1 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr14_-_21979428 | 0.42 |
ENST00000538267.1 ENST00000298717.4 |
METTL3 |
methyltransferase like 3 |
| chr4_-_3534139 | 0.42 |
ENST00000500728.2 |
LRPAP1 |
low density lipoprotein receptor-related protein associated protein 1 |
| chr2_+_37571845 | 0.42 |
ENST00000537448.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr2_+_37571717 | 0.40 |
ENST00000338415.3 ENST00000404976.1 |
QPCT |
glutaminyl-peptide cyclotransferase |
| chr3_-_49941042 | 0.40 |
ENST00000344206.4 ENST00000296474.3 |
MST1R |
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr17_-_1389419 | 0.40 |
ENST00000575158.1 |
MYO1C |
myosin IC |
| chr22_-_45608324 | 0.40 |
ENST00000496226.1 ENST00000251993.7 |
KIAA0930 |
KIAA0930 |
| chr16_+_29984962 | 0.40 |
ENST00000308893.4 |
TAOK2 |
TAO kinase 2 |
| chr6_-_46459675 | 0.39 |
ENST00000306764.7 |
RCAN2 |
regulator of calcineurin 2 |
| chr6_+_35310312 | 0.38 |
ENST00000448077.2 ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD |
peroxisome proliferator-activated receptor delta |
| chr1_+_44445549 | 0.38 |
ENST00000356836.6 |
B4GALT2 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr9_-_113800317 | 0.38 |
ENST00000374431.3 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr13_+_38923959 | 0.38 |
ENST00000379649.1 ENST00000239878.4 ENST00000437952.1 ENST00000379641.1 |
UFM1 |
ubiquitin-fold modifier 1 |
| chr14_+_94492674 | 0.38 |
ENST00000203664.5 ENST00000553723.1 |
OTUB2 |
OTU domain, ubiquitin aldehyde binding 2 |
| chr9_+_35538616 | 0.38 |
ENST00000455600.1 |
RUSC2 |
RUN and SH3 domain containing 2 |
| chr3_-_49722523 | 0.38 |
ENST00000448220.1 |
MST1 |
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr3_+_184032283 | 0.38 |
ENST00000346169.2 ENST00000414031.1 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr7_+_73868120 | 0.37 |
ENST00000265755.3 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
| chr2_-_20251744 | 0.37 |
ENST00000175091.4 |
LAPTM4A |
lysosomal protein transmembrane 4 alpha |
| chr12_-_110434183 | 0.37 |
ENST00000360185.4 ENST00000354574.4 ENST00000338373.5 ENST00000343646.5 ENST00000356259.4 ENST00000553118.1 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr3_+_139654018 | 0.37 |
ENST00000458420.3 |
CLSTN2 |
calsyntenin 2 |
| chr19_-_36297348 | 0.37 |
ENST00000589835.1 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chr1_-_203055129 | 0.37 |
ENST00000241651.4 |
MYOG |
myogenin (myogenic factor 4) |
| chr1_-_98386543 | 0.37 |
ENST00000423006.2 ENST00000370192.3 ENST00000306031.5 |
DPYD |
dihydropyrimidine dehydrogenase |
| chr17_+_39969183 | 0.37 |
ENST00000321562.4 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr2_+_203241531 | 0.37 |
ENST00000374580.4 |
BMPR2 |
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
| chr20_-_36156293 | 0.37 |
ENST00000373537.2 ENST00000414542.2 |
BLCAP |
bladder cancer associated protein |
| chr17_-_1389228 | 0.36 |
ENST00000438665.2 |
MYO1C |
myosin IC |
| chr14_+_96505659 | 0.36 |
ENST00000555004.1 |
C14orf132 |
chromosome 14 open reading frame 132 |
| chr17_+_72744791 | 0.36 |
ENST00000583369.1 ENST00000262613.5 |
SLC9A3R1 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
| chr1_+_40506392 | 0.35 |
ENST00000414893.1 ENST00000414281.1 ENST00000420216.1 ENST00000372792.2 ENST00000372798.1 ENST00000340450.3 ENST00000372805.3 ENST00000435719.1 ENST00000427843.1 ENST00000417287.1 ENST00000424977.1 ENST00000446031.1 |
CAP1 |
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr17_+_46125707 | 0.35 |
ENST00000584137.1 ENST00000362042.3 ENST00000585291.1 ENST00000357480.5 |
NFE2L1 |
nuclear factor, erythroid 2-like 1 |
| chr10_-_32217717 | 0.35 |
ENST00000396144.4 ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12 |
Rho GTPase activating protein 12 |
| chr20_-_36156125 | 0.35 |
ENST00000397135.1 ENST00000397137.1 |
BLCAP |
bladder cancer associated protein |
| chr3_+_184032919 | 0.35 |
ENST00000427845.1 ENST00000342981.4 ENST00000319274.6 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr6_+_89790490 | 0.34 |
ENST00000336032.3 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr2_+_239335449 | 0.34 |
ENST00000264607.4 |
ASB1 |
ankyrin repeat and SOCS box containing 1 |
| chr2_+_85132749 | 0.34 |
ENST00000233143.4 |
TMSB10 |
thymosin beta 10 |
| chr14_-_91526922 | 0.34 |
ENST00000418736.2 ENST00000261991.3 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr6_-_35464727 | 0.33 |
ENST00000402886.3 |
TEAD3 |
TEA domain family member 3 |
| chr3_-_133969437 | 0.33 |
ENST00000460933.1 ENST00000296084.4 |
RYK |
receptor-like tyrosine kinase |
| chr2_-_165477971 | 0.33 |
ENST00000446413.2 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr19_+_7600584 | 0.32 |
ENST00000600737.1 |
PNPLA6 |
patatin-like phospholipase domain containing 6 |
| chr1_+_178062855 | 0.30 |
ENST00000448150.3 |
RASAL2 |
RAS protein activator like 2 |
| chr14_-_103523745 | 0.30 |
ENST00000361246.2 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
| chr22_+_24115000 | 0.30 |
ENST00000215743.3 |
MMP11 |
matrix metallopeptidase 11 (stromelysin 3) |
| chr17_-_2614927 | 0.30 |
ENST00000435359.1 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
| chr7_+_144052381 | 0.30 |
ENST00000498580.1 ENST00000056217.5 |
ARHGEF5 |
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr6_-_35464817 | 0.29 |
ENST00000338863.7 |
TEAD3 |
TEA domain family member 3 |
| chr9_+_32384617 | 0.29 |
ENST00000379923.1 ENST00000309951.6 ENST00000541043.1 |
ACO1 |
aconitase 1, soluble |
| chr6_+_30850697 | 0.29 |
ENST00000509639.1 ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr6_+_35310391 | 0.29 |
ENST00000337400.2 ENST00000311565.4 ENST00000540939.1 |
PPARD |
peroxisome proliferator-activated receptor delta |
| chr11_+_46402297 | 0.28 |
ENST00000405308.2 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr12_-_110434021 | 0.28 |
ENST00000355312.3 ENST00000551209.1 ENST00000550186.1 |
GIT2 |
G protein-coupled receptor kinase interacting ArfGAP 2 |
| chr14_-_77787198 | 0.28 |
ENST00000261534.4 |
POMT2 |
protein-O-mannosyltransferase 2 |
| chr19_-_15311713 | 0.27 |
ENST00000601011.1 ENST00000263388.2 |
NOTCH3 |
notch 3 |
| chr2_-_47168906 | 0.27 |
ENST00000444761.2 ENST00000409147.1 |
MCFD2 |
multiple coagulation factor deficiency 2 |
| chr19_+_797443 | 0.27 |
ENST00000394601.4 ENST00000589575.1 |
PTBP1 |
polypyrimidine tract binding protein 1 |
| chr1_+_40505891 | 0.27 |
ENST00000372797.3 ENST00000372802.1 ENST00000449311.1 |
CAP1 |
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr22_+_45098067 | 0.27 |
ENST00000336985.6 ENST00000403696.1 ENST00000457960.1 ENST00000361473.5 |
PRR5 PRR5-ARHGAP8 |
proline rich 5 (renal) PRR5-ARHGAP8 readthrough |
| chr11_-_33795893 | 0.27 |
ENST00000526785.1 ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3 |
F-box protein 3 |
| chr10_+_99258625 | 0.26 |
ENST00000370664.3 |
UBTD1 |
ubiquitin domain containing 1 |
| chr9_-_100935043 | 0.26 |
ENST00000343933.5 |
CORO2A |
coronin, actin binding protein, 2A |
| chr11_-_18343669 | 0.26 |
ENST00000396253.3 ENST00000349215.3 ENST00000438420.2 |
HPS5 |
Hermansky-Pudlak syndrome 5 |
| chr17_+_61851157 | 0.26 |
ENST00000578681.1 ENST00000583590.1 |
DDX42 |
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr15_-_89456593 | 0.26 |
ENST00000558029.1 ENST00000539437.1 ENST00000542878.1 ENST00000268151.7 ENST00000566497.1 |
MFGE8 |
milk fat globule-EGF factor 8 protein |
| chr7_+_100273736 | 0.25 |
ENST00000412215.1 ENST00000393924.1 |
GNB2 |
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr17_+_79008940 | 0.25 |
ENST00000392411.3 ENST00000575989.1 ENST00000321280.7 ENST00000428708.2 ENST00000575712.1 ENST00000575245.1 ENST00000435091.3 ENST00000321300.6 |
BAIAP2 |
BAI1-associated protein 2 |
| chr17_+_1945301 | 0.25 |
ENST00000572195.1 |
OVCA2 |
ovarian tumor suppressor candidate 2 |
| chrX_+_95939638 | 0.25 |
ENST00000373061.3 ENST00000373054.4 ENST00000355827.4 |
DIAPH2 |
diaphanous-related formin 2 |
| chr1_+_202995611 | 0.24 |
ENST00000367240.2 |
PPFIA4 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 |
| chr3_-_49131013 | 0.24 |
ENST00000424300.1 |
QRICH1 |
glutamine-rich 1 |
| chr12_+_120933859 | 0.24 |
ENST00000242577.6 ENST00000548214.1 ENST00000392508.2 |
DYNLL1 |
dynein, light chain, LC8-type 1 |
| chr1_+_203096831 | 0.24 |
ENST00000337894.4 |
ADORA1 |
adenosine A1 receptor |
| chr12_+_130646999 | 0.24 |
ENST00000539839.1 ENST00000229030.4 |
FZD10 |
frizzled family receptor 10 |
| chr9_+_114423615 | 0.23 |
ENST00000374293.4 |
GNG10 |
guanine nucleotide binding protein (G protein), gamma 10 |
| chr19_+_41725140 | 0.23 |
ENST00000359092.3 |
AXL |
AXL receptor tyrosine kinase |
| chr6_-_83902933 | 0.23 |
ENST00000512866.1 ENST00000510258.1 ENST00000503094.1 ENST00000283977.4 ENST00000513973.1 ENST00000508748.1 |
PGM3 |
phosphoglucomutase 3 |
| chr9_-_113800341 | 0.23 |
ENST00000358883.4 |
LPAR1 |
lysophosphatidic acid receptor 1 |
| chr16_+_58035277 | 0.23 |
ENST00000219281.3 ENST00000539737.2 ENST00000423271.3 ENST00000561568.1 ENST00000563149.1 |
USB1 |
U6 snRNA biogenesis 1 |
| chr5_-_180237445 | 0.23 |
ENST00000393340.3 |
MGAT1 |
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr9_+_35732312 | 0.23 |
ENST00000353704.2 |
CREB3 |
cAMP responsive element binding protein 3 |
| chr19_-_42192189 | 0.22 |
ENST00000401731.1 ENST00000338196.4 ENST00000006724.3 |
CEACAM7 |
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr1_+_15250596 | 0.22 |
ENST00000361144.5 |
KAZN |
kazrin, periplakin interacting protein |
| chr1_+_203764742 | 0.22 |
ENST00000432282.1 ENST00000453771.1 ENST00000367214.1 ENST00000367212.3 ENST00000332127.4 |
ZC3H11A |
zinc finger CCCH-type containing 11A |
| chr3_-_88108212 | 0.22 |
ENST00000482016.1 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr2_+_28974603 | 0.22 |
ENST00000441461.1 ENST00000358506.2 |
PPP1CB |
protein phosphatase 1, catalytic subunit, beta isozyme |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 1.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.0 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.8 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 2.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.9 | 2.7 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.4 | 1.6 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.3 | 1.4 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.3 | 1.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 1.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.2 | 0.6 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.2 | 1.0 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.4 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.4 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 1.4 | GO:0033643 | host cell part(GO:0033643) |
| 0.1 | 0.5 | GO:1990130 | EGO complex(GO:0034448) Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.4 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 1.0 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.5 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 0.7 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.4 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 1.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.1 | 6.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.1 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 1.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 2.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.8 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.9 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.9 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 3.3 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 1.4 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 7.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 1.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 1.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.7 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.6 | 1.9 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.6 | 8.4 | GO:0035878 | nail development(GO:0035878) |
| 0.6 | 1.7 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.5 | 4.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.5 | 4.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.5 | 3.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.4 | 2.6 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.3 | 1.4 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.3 | 1.0 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.3 | 0.9 | GO:0048627 | myoblast development(GO:0048627) |
| 0.3 | 0.9 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.3 | 1.8 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.3 | 2.4 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.3 | 1.1 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.3 | 0.8 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.3 | 0.8 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.3 | 1.6 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.3 | 1.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.2 | 1.0 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 0.9 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 1.1 | GO:0030047 | actin modification(GO:0030047) |
| 0.2 | 1.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 1.7 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.2 | 0.6 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.2 | 0.5 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.2 | 0.3 | GO:0061643 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.2 | 0.6 | GO:0032811 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.2 | 0.6 | GO:1904566 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 1.6 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.1 | 0.7 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.8 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.9 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.5 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.5 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.1 | 1.5 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.4 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 0.4 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.4 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.4 | GO:0003250 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.1 | 1.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.6 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.1 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.6 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 0.8 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.1 | 0.4 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.6 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.9 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.1 | 0.8 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.7 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 0.6 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.4 | GO:0032416 | negative regulation of sodium:proton antiporter activity(GO:0032416) |
| 0.1 | 0.4 | GO:1904098 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.1 | 0.3 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 1.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.9 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.3 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) histone H2A phosphorylation(GO:1990164) |
| 0.1 | 1.0 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) positive regulation of membrane potential(GO:0045838) |
| 0.1 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.2 | GO:0032242 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) |
| 0.1 | 0.4 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 1.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.0 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.3 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.2 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.2 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.3 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.1 | 1.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 1.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.4 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.3 | GO:0072104 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.5 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.4 | GO:1900223 | positive regulation of beta-amyloid clearance(GO:1900223) |
| 0.0 | 0.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.4 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.5 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.5 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.4 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.0 | 0.3 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0009227 | UDP-glucose catabolic process(GO:0006258) nucleotide-sugar catabolic process(GO:0009227) |
| 0.0 | 0.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0051586 | peptidyl-cysteine methylation(GO:0018125) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 1.4 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 1.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0032596 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 1.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 1.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.7 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 1.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 1.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.3 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.4 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.7 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.0 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 1.8 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.2 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.3 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.1 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.5 | GO:0018146 | keratan sulfate biosynthetic process(GO:0018146) |
| 0.0 | 0.3 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.4 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.3 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 0.5 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.4 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.5 | 2.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.3 | 1.4 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.3 | 1.3 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.3 | 0.9 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.3 | 0.8 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.2 | 0.6 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 3.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 0.8 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.2 | 1.6 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 1.5 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.2 | 4.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 2.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 1.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.5 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.4 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.4 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.1 | 0.5 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 0.9 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.5 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 4.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 3.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.9 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.7 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.3 | GO:0051538 | iron-responsive element binding(GO:0030350) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 1.4 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 0.9 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.4 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 1.8 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 1.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 1.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.6 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.3 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.8 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 1.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.2 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 2.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0030305 | heparanase activity(GO:0030305) |
| 0.0 | 1.0 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 1.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.4 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 4.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.7 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 1.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 1.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 2.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 3.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 1.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.9 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 4.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 4.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 8.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 2.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.0 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |