Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
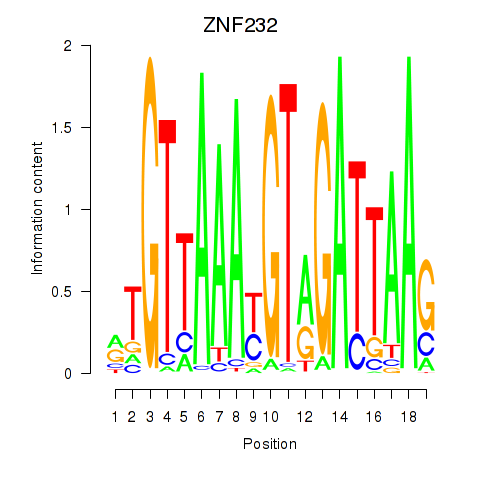
Results for ZNF232
Z-value: 0.47
Transcription factors associated with ZNF232
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF232
|
ENSG00000167840.9 | ZNF232 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF232 | hg19_v2_chr17_-_5015129_5015187 | -0.03 | 9.3e-01 | Click! |
Activity profile of ZNF232 motif
Sorted Z-values of ZNF232 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF232
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_117085336 | 1.12 |
ENST00000259396.8 ENST00000538816.1 |
ORM1 |
orosomucoid 1 |
| chrY_+_15016725 | 1.11 |
ENST00000336079.3 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chrX_-_130423386 | 1.06 |
ENST00000370903.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr18_+_55816546 | 0.86 |
ENST00000435432.2 ENST00000357895.5 ENST00000586263.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr3_+_8543393 | 0.75 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr2_+_102413726 | 0.72 |
ENST00000350878.4 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr1_-_79472365 | 0.66 |
ENST00000370742.3 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
| chrY_+_15016013 | 0.63 |
ENST00000360160.4 ENST00000454054.1 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr3_+_8543561 | 0.52 |
ENST00000397386.3 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr6_-_46922659 | 0.42 |
ENST00000265417.7 |
GPR116 |
G protein-coupled receptor 116 |
| chr17_+_39975455 | 0.40 |
ENST00000455106.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr2_+_241807870 | 0.39 |
ENST00000307503.3 |
AGXT |
alanine-glyoxylate aminotransferase |
| chr17_+_39975544 | 0.36 |
ENST00000544340.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr20_-_33732952 | 0.34 |
ENST00000541621.1 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr9_-_115480303 | 0.33 |
ENST00000374234.1 ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP |
INTS3 and NABP interacting protein |
| chr3_+_8543533 | 0.27 |
ENST00000454244.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr2_-_113522177 | 0.23 |
ENST00000541405.1 |
CKAP2L |
cytoskeleton associated protein 2-like |
| chr6_+_109416313 | 0.23 |
ENST00000521277.1 ENST00000517392.1 ENST00000407272.1 ENST00000336977.4 ENST00000519286.1 ENST00000518329.1 ENST00000522461.1 ENST00000518853.1 |
CEP57L1 |
centrosomal protein 57kDa-like 1 |
| chr2_-_74619152 | 0.21 |
ENST00000440727.1 ENST00000409240.1 |
DCTN1 |
dynactin 1 |
| chr3_-_185216811 | 0.20 |
ENST00000421852.1 |
TMEM41A |
transmembrane protein 41A |
| chr11_+_66276550 | 0.20 |
ENST00000419755.3 |
CTD-3074O7.11 |
Bardet-Biedl syndrome 1 protein |
| chr2_-_74618964 | 0.18 |
ENST00000417090.1 ENST00000409868.1 |
DCTN1 |
dynactin 1 |
| chr20_+_4129426 | 0.18 |
ENST00000339123.6 ENST00000305958.4 ENST00000278795.3 |
SMOX |
spermine oxidase |
| chr10_-_115904361 | 0.16 |
ENST00000428953.1 ENST00000543782.1 |
C10orf118 |
chromosome 10 open reading frame 118 |
| chr3_-_123304017 | 0.15 |
ENST00000383657.5 |
PTPLB |
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member b |
| chr3_-_185216766 | 0.15 |
ENST00000296254.3 |
TMEM41A |
transmembrane protein 41A |
| chr17_-_6915646 | 0.14 |
ENST00000574377.1 ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2 |
Uncharacterized protein |
| chr16_+_30662360 | 0.14 |
ENST00000542965.2 |
PRR14 |
proline rich 14 |
| chrX_+_49832231 | 0.14 |
ENST00000376108.3 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
| chr2_-_113522248 | 0.13 |
ENST00000302450.6 |
CKAP2L |
cytoskeleton associated protein 2-like |
| chr5_-_180076613 | 0.12 |
ENST00000261937.6 ENST00000393347.3 |
FLT4 |
fms-related tyrosine kinase 4 |
| chrX_-_100548045 | 0.10 |
ENST00000372907.3 ENST00000372905.2 |
TAF7L |
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
| chr16_-_4852915 | 0.09 |
ENST00000322048.7 |
ROGDI |
rogdi homolog (Drosophila) |
| chr19_-_10491234 | 0.08 |
ENST00000524462.1 ENST00000531836.1 ENST00000525621.1 |
TYK2 |
tyrosine kinase 2 |
| chr5_-_180076580 | 0.07 |
ENST00000502649.1 |
FLT4 |
fms-related tyrosine kinase 4 |
| chr16_+_30662184 | 0.07 |
ENST00000300835.4 |
PRR14 |
proline rich 14 |
| chr14_-_24711806 | 0.06 |
ENST00000540705.1 ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr15_-_79383102 | 0.06 |
ENST00000558480.2 ENST00000419573.3 |
RASGRF1 |
Ras protein-specific guanine nucleotide-releasing factor 1 |
| chr14_-_24711865 | 0.05 |
ENST00000399423.4 ENST00000267415.7 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr1_-_53686261 | 0.03 |
ENST00000294360.4 |
C1orf123 |
chromosome 1 open reading frame 123 |
| chr22_-_30234218 | 0.01 |
ENST00000307790.3 ENST00000542393.1 ENST00000397771.2 |
ASCC2 |
activating signal cointegrator 1 complex subunit 2 |
| chr21_+_38792602 | 0.01 |
ENST00000398960.2 ENST00000398956.2 |
DYRK1A |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chrX_+_100805496 | 0.01 |
ENST00000372829.3 |
ARMCX1 |
armadillo repeat containing, X-linked 1 |
| chr15_-_56757329 | 0.01 |
ENST00000260453.3 |
MNS1 |
meiosis-specific nuclear structural 1 |
| chr8_-_143999259 | 0.00 |
ENST00000323110.2 |
CYP11B2 |
cytochrome P450, family 11, subfamily B, polypeptide 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.2 | 0.9 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.1 | GO:1904469 | positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.1 | 0.4 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.1 | 0.3 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 1.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.4 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 1.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.1 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.2 | GO:0060312 | regulation of blood vessel remodeling(GO:0060312) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.2 | GO:0052894 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.7 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.9 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.1 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.3 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |