Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
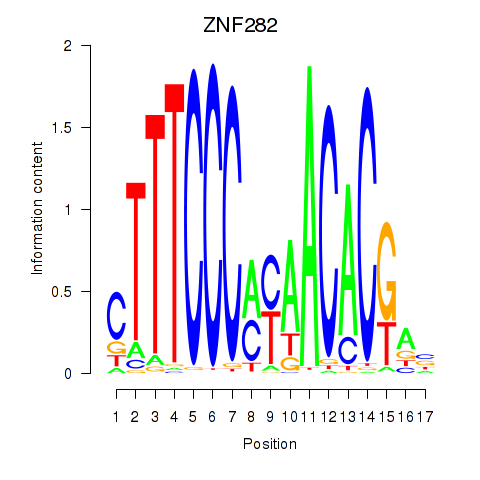
Results for ZNF282
Z-value: 0.44
Transcription factors associated with ZNF282
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF282
|
ENSG00000170265.7 | ZNF282 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF282 | hg19_v2_chr7_+_148892557_148892580 | -0.40 | 1.3e-01 | Click! |
Activity profile of ZNF282 motif
Sorted Z-values of ZNF282 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF282
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_169753156 | 0.89 |
ENST00000393726.3 ENST00000507735.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr6_+_106959718 | 0.81 |
ENST00000369066.3 |
AIM1 |
absent in melanoma 1 |
| chrX_-_13835147 | 0.58 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr17_+_42422662 | 0.52 |
ENST00000593167.1 ENST00000585512.1 ENST00000591740.1 ENST00000592783.1 ENST00000587387.1 ENST00000588237.1 ENST00000589265.1 |
GRN |
granulin |
| chr17_+_42422637 | 0.51 |
ENST00000053867.3 ENST00000588143.1 |
GRN |
granulin |
| chr20_+_18269121 | 0.46 |
ENST00000377671.3 ENST00000360010.5 ENST00000396026.3 ENST00000402618.2 ENST00000401790.1 ENST00000434018.1 ENST00000538547.1 ENST00000535822.1 |
ZNF133 |
zinc finger protein 133 |
| chr22_+_45898712 | 0.44 |
ENST00000455233.1 ENST00000348697.2 ENST00000402984.3 ENST00000262722.7 ENST00000327858.6 ENST00000442170.2 ENST00000340923.5 ENST00000439835.1 |
FBLN1 |
fibulin 1 |
| chr6_+_129204337 | 0.44 |
ENST00000421865.2 |
LAMA2 |
laminin, alpha 2 |
| chr12_-_122296755 | 0.41 |
ENST00000289004.4 |
HPD |
4-hydroxyphenylpyruvate dioxygenase |
| chr17_+_7942335 | 0.37 |
ENST00000380183.4 ENST00000572022.1 ENST00000380173.2 |
ALOX15B |
arachidonate 15-lipoxygenase, type B |
| chr14_+_55590646 | 0.33 |
ENST00000553493.1 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
| chr10_+_114135004 | 0.32 |
ENST00000393081.1 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
| chr1_+_2004901 | 0.30 |
ENST00000400921.2 |
PRKCZ |
protein kinase C, zeta |
| chrX_-_13835461 | 0.30 |
ENST00000316715.4 ENST00000356942.5 |
GPM6B |
glycoprotein M6B |
| chr21_+_35747773 | 0.29 |
ENST00000399292.3 ENST00000399299.1 ENST00000399295.2 |
SMIM11 |
small integral membrane protein 11 |
| chr17_+_66509019 | 0.27 |
ENST00000585981.1 ENST00000589480.1 ENST00000585815.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr2_+_219283815 | 0.24 |
ENST00000248444.5 ENST00000454069.1 ENST00000392114.2 |
VIL1 |
villin 1 |
| chr1_+_35247859 | 0.22 |
ENST00000373362.3 |
GJB3 |
gap junction protein, beta 3, 31kDa |
| chr8_-_23261589 | 0.22 |
ENST00000524168.1 ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2 |
lysyl oxidase-like 2 |
| chr4_-_153457197 | 0.22 |
ENST00000281708.4 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr16_+_67204400 | 0.22 |
ENST00000563439.1 ENST00000432069.2 ENST00000564992.1 ENST00000564053.1 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr13_+_31309645 | 0.22 |
ENST00000380490.3 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
| chr11_+_66278080 | 0.22 |
ENST00000318312.7 ENST00000526815.1 ENST00000537537.1 ENST00000525809.1 ENST00000455748.2 ENST00000393994.2 |
BBS1 |
Bardet-Biedl syndrome 1 |
| chr2_+_242289502 | 0.21 |
ENST00000451310.1 |
SEPT2 |
septin 2 |
| chr19_-_39390440 | 0.21 |
ENST00000249396.7 ENST00000414941.1 ENST00000392081.2 |
SIRT2 |
sirtuin 2 |
| chr9_+_126777676 | 0.20 |
ENST00000488674.2 |
LHX2 |
LIM homeobox 2 |
| chr1_+_2005425 | 0.20 |
ENST00000461106.2 |
PRKCZ |
protein kinase C, zeta |
| chr20_-_43150601 | 0.20 |
ENST00000541235.1 ENST00000255175.1 ENST00000342374.4 |
SERINC3 |
serine incorporator 3 |
| chr1_-_9129735 | 0.19 |
ENST00000377424.4 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr5_+_78365577 | 0.18 |
ENST00000518666.1 ENST00000521567.1 |
BHMT2 |
betaine--homocysteine S-methyltransferase 2 |
| chr1_-_24151892 | 0.18 |
ENST00000235958.4 |
HMGCL |
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr1_+_13910479 | 0.17 |
ENST00000509009.1 |
PDPN |
podoplanin |
| chr1_+_145507587 | 0.17 |
ENST00000330165.8 ENST00000369307.3 |
RBM8A |
RNA binding motif protein 8A |
| chr1_-_24151903 | 0.17 |
ENST00000436439.2 ENST00000374490.3 |
HMGCL |
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr1_+_151171012 | 0.16 |
ENST00000349792.5 ENST00000409426.1 ENST00000441902.2 ENST00000368890.4 ENST00000424999.1 ENST00000368888.4 |
PIP5K1A |
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
| chr19_-_39390350 | 0.16 |
ENST00000447739.1 ENST00000358931.5 ENST00000407552.1 |
SIRT2 |
sirtuin 2 |
| chr1_-_13390765 | 0.15 |
ENST00000357367.2 |
PRAMEF8 |
PRAME family member 8 |
| chr3_-_106959424 | 0.15 |
ENST00000607801.1 ENST00000479612.2 ENST00000484698.1 ENST00000477210.2 ENST00000473636.1 |
LINC00882 |
long intergenic non-protein coding RNA 882 |
| chr1_+_160313062 | 0.15 |
ENST00000294785.5 ENST00000368063.1 ENST00000437169.1 |
NCSTN |
nicastrin |
| chr12_-_46766577 | 0.15 |
ENST00000256689.5 |
SLC38A2 |
solute carrier family 38, member 2 |
| chr1_+_13910757 | 0.14 |
ENST00000376061.4 ENST00000513143.1 |
PDPN |
podoplanin |
| chr5_+_175299743 | 0.12 |
ENST00000502265.1 |
CPLX2 |
complexin 2 |
| chr16_+_67927147 | 0.12 |
ENST00000291041.5 |
PSKH1 |
protein serine kinase H1 |
| chr16_-_70239683 | 0.12 |
ENST00000601706.1 |
AC009060.1 |
Uncharacterized protein |
| chr17_-_77924627 | 0.11 |
ENST00000572862.1 ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16 |
TBC1 domain family, member 16 |
| chr5_+_167718604 | 0.11 |
ENST00000265293.4 |
WWC1 |
WW and C2 domain containing 1 |
| chr10_+_95753714 | 0.11 |
ENST00000260766.3 |
PLCE1 |
phospholipase C, epsilon 1 |
| chr19_-_58220517 | 0.11 |
ENST00000512439.2 ENST00000426889.1 |
ZNF154 |
zinc finger protein 154 |
| chr1_-_9129631 | 0.11 |
ENST00000377414.3 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr2_-_9695847 | 0.10 |
ENST00000310823.3 ENST00000497134.1 |
ADAM17 |
ADAM metallopeptidase domain 17 |
| chr6_+_39760783 | 0.10 |
ENST00000398904.2 ENST00000538976.1 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr7_+_139025875 | 0.09 |
ENST00000297534.6 |
C7orf55 |
chromosome 7 open reading frame 55 |
| chr18_+_54318566 | 0.09 |
ENST00000589935.1 ENST00000357574.3 |
WDR7 |
WD repeat domain 7 |
| chr1_+_17248418 | 0.09 |
ENST00000375541.5 |
CROCC |
ciliary rootlet coiled-coil, rootletin |
| chr19_+_44617511 | 0.08 |
ENST00000262894.6 ENST00000588926.1 ENST00000592780.1 |
ZNF225 |
zinc finger protein 225 |
| chr12_-_9102549 | 0.08 |
ENST00000000412.3 |
M6PR |
mannose-6-phosphate receptor (cation dependent) |
| chr15_+_51200871 | 0.08 |
ENST00000560508.1 |
AP4E1 |
adaptor-related protein complex 4, epsilon 1 subunit |
| chr7_+_16828866 | 0.08 |
ENST00000597084.1 |
AC073333.1 |
Uncharacterized protein |
| chr5_-_175788758 | 0.08 |
ENST00000510164.1 ENST00000533553.1 ENST00000504688.1 ENST00000393725.2 ENST00000503082.1 ENST00000506983.1 |
KIAA1191 |
KIAA1191 |
| chr5_+_140552218 | 0.06 |
ENST00000231137.3 |
PCDHB7 |
protocadherin beta 7 |
| chr17_+_41323204 | 0.06 |
ENST00000542611.1 ENST00000590996.1 ENST00000389312.4 ENST00000589872.1 |
NBR1 |
neighbor of BRCA1 gene 1 |
| chr15_-_44955842 | 0.06 |
ENST00000427534.2 ENST00000559193.1 ENST00000261866.7 ENST00000535302.2 ENST00000558319.1 |
SPG11 |
spastic paraplegia 11 (autosomal recessive) |
| chr17_-_56605341 | 0.06 |
ENST00000583114.1 |
SEPT4 |
septin 4 |
| chr19_+_7953384 | 0.06 |
ENST00000306708.6 |
LRRC8E |
leucine rich repeat containing 8 family, member E |
| chr2_+_233415363 | 0.06 |
ENST00000409514.1 ENST00000409098.1 ENST00000409495.1 ENST00000409167.3 ENST00000409322.1 ENST00000409394.1 |
EIF4E2 |
eukaryotic translation initiation factor 4E family member 2 |
| chr12_-_13248562 | 0.05 |
ENST00000457134.2 ENST00000537302.1 |
GSG1 |
germ cell associated 1 |
| chr6_+_32938665 | 0.05 |
ENST00000374831.4 ENST00000395289.2 |
BRD2 |
bromodomain containing 2 |
| chr18_-_54318353 | 0.05 |
ENST00000590954.1 ENST00000540155.1 |
TXNL1 |
thioredoxin-like 1 |
| chr9_+_34652164 | 0.04 |
ENST00000441545.2 ENST00000553620.1 |
IL11RA |
interleukin 11 receptor, alpha |
| chr17_+_44039704 | 0.04 |
ENST00000420682.2 ENST00000415613.2 ENST00000571987.1 ENST00000574436.1 ENST00000431008.3 |
MAPT |
microtubule-associated protein tau |
| chr1_-_155947951 | 0.04 |
ENST00000313695.7 ENST00000497907.1 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr3_+_45927994 | 0.04 |
ENST00000357632.2 ENST00000395963.2 |
CCR9 |
chemokine (C-C motif) receptor 9 |
| chr6_-_79944336 | 0.04 |
ENST00000344726.5 ENST00000275036.7 |
HMGN3 |
high mobility group nucleosomal binding domain 3 |
| chr14_-_24911448 | 0.04 |
ENST00000555355.1 ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
| chr15_+_51200859 | 0.04 |
ENST00000261842.5 |
AP4E1 |
adaptor-related protein complex 4, epsilon 1 subunit |
| chr1_+_52682052 | 0.03 |
ENST00000371591.1 |
ZFYVE9 |
zinc finger, FYVE domain containing 9 |
| chr12_-_49582978 | 0.03 |
ENST00000301071.7 |
TUBA1A |
tubulin, alpha 1a |
| chr12_-_13248598 | 0.03 |
ENST00000337630.6 ENST00000545699.1 |
GSG1 |
germ cell associated 1 |
| chr19_-_46285736 | 0.03 |
ENST00000291270.4 ENST00000447742.2 ENST00000354227.5 |
DMPK |
dystrophia myotonica-protein kinase |
| chr11_-_66445219 | 0.03 |
ENST00000525754.1 ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B |
RNA binding motif protein 4B |
| chr16_+_30207122 | 0.03 |
ENST00000395137.2 |
SULT1A3 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr8_-_27630102 | 0.02 |
ENST00000356537.4 ENST00000522915.1 ENST00000539095.1 |
CCDC25 |
coiled-coil domain containing 25 |
| chr19_-_51340469 | 0.02 |
ENST00000326856.4 |
KLK15 |
kallikrein-related peptidase 15 |
| chr16_+_29467780 | 0.02 |
ENST00000395400.3 |
SULT1A4 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4 |
| chr11_-_6633799 | 0.01 |
ENST00000299424.4 |
TAF10 |
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr20_-_2451395 | 0.01 |
ENST00000339610.6 ENST00000381342.2 ENST00000438552.2 |
SNRPB |
small nuclear ribonucleoprotein polypeptides B and B1 |
| chr5_+_65892174 | 0.01 |
ENST00000404260.3 ENST00000403625.2 ENST00000406374.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr19_+_39390587 | 0.01 |
ENST00000572515.1 ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr2_-_68479614 | 0.00 |
ENST00000234310.3 |
PPP3R1 |
protein phosphatase 3, regulatory subunit B, alpha |
| chr19_-_6110474 | 0.00 |
ENST00000587181.1 ENST00000587321.1 ENST00000586806.1 ENST00000589742.1 ENST00000592546.1 ENST00000303657.5 |
RFX2 |
regulatory factor X, 2 (influences HLA class II expression) |
| chr18_+_54318616 | 0.00 |
ENST00000254442.3 |
WDR7 |
WD repeat domain 7 |
| chr17_-_73389854 | 0.00 |
ENST00000578961.1 ENST00000392564.1 ENST00000582582.1 |
GRB2 |
growth factor receptor-bound protein 2 |
| chr2_+_233415488 | 0.00 |
ENST00000454501.1 |
EIF4E2 |
eukaryotic translation initiation factor 4E family member 2 |
| chr6_+_32938692 | 0.00 |
ENST00000443797.2 |
BRD2 |
bromodomain containing 2 |
| chr6_+_144471643 | 0.00 |
ENST00000367568.4 |
STX11 |
syntaxin 11 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.4 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.4 | GO:0050473 | arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.1 | 0.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.2 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.1 | 0.2 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 0.3 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.3 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.5 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 0.3 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.1 | 0.3 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.1 | 0.9 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.5 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.2 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 0.4 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.2 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.4 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.3 | GO:1990539 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.9 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.2 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.4 | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading(GO:1900025) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of meiotic nuclear division(GO:0045835) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.2 | GO:0071267 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 0.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.0 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.4 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.4 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.1 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0031588 | cAMP-dependent protein kinase complex(GO:0005952) nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |