Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
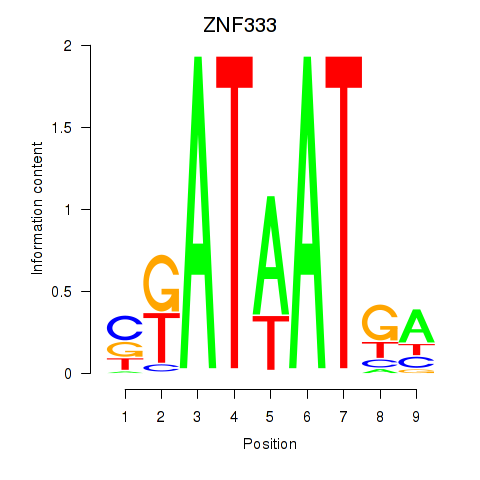
Results for ZNF333
Z-value: 0.62
Transcription factors associated with ZNF333
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF333
|
ENSG00000160961.7 | ZNF333 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF333 | hg19_v2_chr19_+_14800711_14800917 | -0.61 | 1.2e-02 | Click! |
Activity profile of ZNF333 motif
Sorted Z-values of ZNF333 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF333
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_159557637 | 2.10 |
ENST00000445224.2 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr12_-_71031185 | 1.95 |
ENST00000548122.1 ENST00000551525.1 ENST00000550358.1 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chrX_-_21776281 | 1.24 |
ENST00000379494.3 |
SMPX |
small muscle protein, X-linked |
| chr12_-_71031220 | 1.07 |
ENST00000334414.6 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr2_-_106013400 | 1.07 |
ENST00000409807.1 |
FHL2 |
four and a half LIM domains 2 |
| chr10_+_54074033 | 1.00 |
ENST00000373970.3 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
| chr14_+_85996471 | 0.90 |
ENST00000330753.4 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr10_+_45495898 | 0.85 |
ENST00000298299.3 |
ZNF22 |
zinc finger protein 22 |
| chr14_+_85996507 | 0.65 |
ENST00000554746.1 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr3_+_35721106 | 0.65 |
ENST00000474696.1 ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr7_-_115670804 | 0.65 |
ENST00000320239.7 |
TFEC |
transcription factor EC |
| chr7_-_115670792 | 0.61 |
ENST00000265440.7 ENST00000393485.1 |
TFEC |
transcription factor EC |
| chr12_-_10251539 | 0.56 |
ENST00000420265.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr3_+_173116225 | 0.55 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr10_+_121410882 | 0.45 |
ENST00000369085.3 |
BAG3 |
BCL2-associated athanogene 3 |
| chr1_+_11994715 | 0.43 |
ENST00000449038.1 ENST00000376369.3 ENST00000429000.2 ENST00000196061.4 |
PLOD1 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr2_-_145275228 | 0.42 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr2_+_233527443 | 0.42 |
ENST00000410095.1 |
EFHD1 |
EF-hand domain family, member D1 |
| chr1_-_36789755 | 0.41 |
ENST00000270824.1 |
EVA1B |
eva-1 homolog B (C. elegans) |
| chr4_-_186696425 | 0.41 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr5_-_133510456 | 0.36 |
ENST00000520417.1 |
SKP1 |
S-phase kinase-associated protein 1 |
| chr5_+_169010638 | 0.28 |
ENST00000265295.4 ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1 |
spindle apparatus coiled-coil protein 1 |
| chr2_-_128432639 | 0.26 |
ENST00000545738.2 ENST00000355119.4 ENST00000409808.2 |
LIMS2 |
LIM and senescent cell antigen-like domains 2 |
| chr1_-_197036364 | 0.26 |
ENST00000367412.1 |
F13B |
coagulation factor XIII, B polypeptide |
| chr8_-_11725549 | 0.26 |
ENST00000505496.2 ENST00000534636.1 ENST00000524500.1 ENST00000345125.3 ENST00000453527.2 ENST00000527215.2 ENST00000532392.1 ENST00000533455.1 ENST00000534510.1 ENST00000530640.2 ENST00000531089.1 ENST00000532656.2 ENST00000531502.1 ENST00000434271.1 ENST00000353047.6 |
CTSB |
cathepsin B |
| chr7_-_27169801 | 0.22 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr3_-_194393206 | 0.22 |
ENST00000265245.5 |
LSG1 |
large 60S subunit nuclear export GTPase 1 |
| chr11_+_12766583 | 0.21 |
ENST00000361985.2 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr8_+_40010989 | 0.18 |
ENST00000315792.3 |
C8orf4 |
chromosome 8 open reading frame 4 |
| chr1_-_51763661 | 0.17 |
ENST00000530004.1 |
TTC39A |
tetratricopeptide repeat domain 39A |
| chr11_-_6440624 | 0.17 |
ENST00000311051.3 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr6_+_160327974 | 0.17 |
ENST00000252660.4 |
MAS1 |
MAS1 oncogene |
| chr1_+_218458625 | 0.15 |
ENST00000366932.3 |
RRP15 |
ribosomal RNA processing 15 homolog (S. cerevisiae) |
| chr17_-_33446735 | 0.15 |
ENST00000460118.2 ENST00000335858.7 |
RAD51D |
RAD51 paralog D |
| chr11_-_66313699 | 0.14 |
ENST00000526986.1 ENST00000310442.3 |
ZDHHC24 |
zinc finger, DHHC-type containing 24 |
| chr12_-_10959892 | 0.14 |
ENST00000240615.2 |
TAS2R8 |
taste receptor, type 2, member 8 |
| chr12_+_117013656 | 0.13 |
ENST00000556529.1 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
| chr22_-_32651326 | 0.13 |
ENST00000266086.4 |
SLC5A4 |
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr11_-_18062872 | 0.12 |
ENST00000250018.2 |
TPH1 |
tryptophan hydroxylase 1 |
| chr5_+_137203465 | 0.12 |
ENST00000239926.4 |
MYOT |
myotilin |
| chr22_+_22516550 | 0.12 |
ENST00000390284.2 |
IGLV4-60 |
immunoglobulin lambda variable 4-60 |
| chr1_+_55013889 | 0.11 |
ENST00000343744.2 ENST00000371316.3 |
ACOT11 |
acyl-CoA thioesterase 11 |
| chr3_-_37216055 | 0.10 |
ENST00000336686.4 |
LRRFIP2 |
leucine rich repeat (in FLII) interacting protein 2 |
| chr11_+_18433840 | 0.10 |
ENST00000541669.1 ENST00000280704.4 |
LDHC |
lactate dehydrogenase C |
| chr8_+_61429416 | 0.09 |
ENST00000262646.7 ENST00000531289.1 |
RAB2A |
RAB2A, member RAS oncogene family |
| chr5_-_35195338 | 0.09 |
ENST00000509839.1 |
PRLR |
prolactin receptor |
| chr17_-_46690839 | 0.08 |
ENST00000498634.2 |
HOXB8 |
homeobox B8 |
| chr12_+_58176525 | 0.08 |
ENST00000543727.1 ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM |
Ts translation elongation factor, mitochondrial |
| chr4_+_106473768 | 0.07 |
ENST00000265154.2 ENST00000420470.2 |
ARHGEF38 |
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr20_+_56964169 | 0.07 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr16_-_28222797 | 0.07 |
ENST00000569951.1 ENST00000565698.1 |
XPO6 |
exportin 6 |
| chr11_-_71823796 | 0.07 |
ENST00000545680.1 ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr2_-_178753465 | 0.06 |
ENST00000389683.3 |
PDE11A |
phosphodiesterase 11A |
| chr8_+_18248755 | 0.05 |
ENST00000286479.3 |
NAT2 |
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr16_-_20702578 | 0.05 |
ENST00000307493.4 ENST00000219151.4 |
ACSM1 |
acyl-CoA synthetase medium-chain family member 1 |
| chr14_+_23842018 | 0.05 |
ENST00000397242.2 ENST00000329715.2 |
IL25 |
interleukin 25 |
| chr6_+_17110726 | 0.05 |
ENST00000354384.5 |
STMND1 |
stathmin domain containing 1 |
| chr12_-_7656357 | 0.05 |
ENST00000396620.3 ENST00000432237.2 ENST00000359156.4 |
CD163 |
CD163 molecule |
| chr5_-_9630463 | 0.05 |
ENST00000382492.2 |
TAS2R1 |
taste receptor, type 2, member 1 |
| chr11_-_71823715 | 0.04 |
ENST00000545944.1 ENST00000502597.2 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chrY_-_6742068 | 0.04 |
ENST00000215479.5 |
AMELY |
amelogenin, Y-linked |
| chr17_-_3337135 | 0.04 |
ENST00000248384.1 |
OR1E2 |
olfactory receptor, family 1, subfamily E, member 2 |
| chr4_-_100356551 | 0.04 |
ENST00000209665.4 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr5_+_140800638 | 0.03 |
ENST00000398587.2 ENST00000518882.1 |
PCDHGA11 |
protocadherin gamma subfamily A, 11 |
| chr12_+_60058458 | 0.03 |
ENST00000548610.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr3_+_38323785 | 0.03 |
ENST00000466887.1 ENST00000448498.1 |
SLC22A14 |
solute carrier family 22, member 14 |
| chr18_+_46065393 | 0.02 |
ENST00000256413.3 |
CTIF |
CBP80/20-dependent translation initiation factor |
| chr13_-_103719196 | 0.02 |
ENST00000245312.3 |
SLC10A2 |
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr6_-_135375986 | 0.02 |
ENST00000525067.1 ENST00000367822.5 ENST00000367837.5 |
HBS1L |
HBS1-like (S. cerevisiae) |
| chr18_+_71815743 | 0.02 |
ENST00000169551.6 ENST00000580087.1 |
TIMM21 |
translocase of inner mitochondrial membrane 21 homolog (yeast) |
| chr4_+_71108300 | 0.01 |
ENST00000304954.3 |
CSN3 |
casein kappa |
| chr2_-_228497888 | 0.01 |
ENST00000264387.4 ENST00000409066.1 |
C2orf83 |
chromosome 2 open reading frame 83 |
| chr3_-_126327398 | 0.01 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr19_-_45004556 | 0.01 |
ENST00000587047.1 ENST00000391956.4 ENST00000221327.4 ENST00000586637.1 ENST00000591064.1 ENST00000592529.1 |
ZNF180 |
zinc finger protein 180 |
| chr1_+_225600404 | 0.01 |
ENST00000366845.2 |
AC092811.1 |
AC092811.1 |
| chr9_-_19786926 | 0.00 |
ENST00000341998.2 ENST00000286344.3 |
SLC24A2 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chrX_+_11311533 | 0.00 |
ENST00000380714.3 ENST00000380712.3 ENST00000348912.4 |
AMELX |
amelogenin, X-linked |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.4 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 3.0 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.1 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.1 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) negative regulation of cell fate specification(GO:0009996) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.3 | 1.6 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 | 1.1 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.2 | 0.5 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.1 | 0.4 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.2 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.0 | 0.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.2 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 3.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.1 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.0 | 0.0 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.4 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 1.9 | GO:0034605 | cellular response to heat(GO:0034605) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 3.0 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.1 | 1.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 1.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0004459 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 1.1 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |