Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
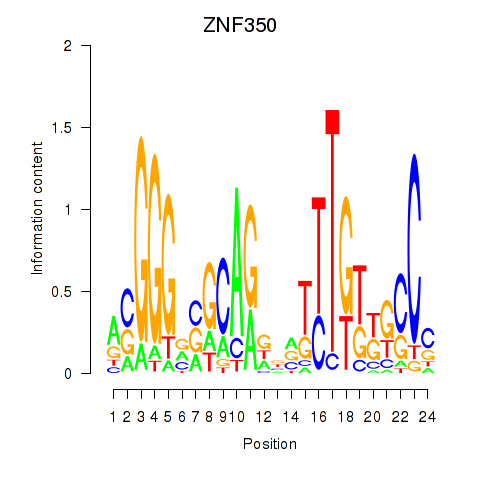
Results for ZNF350
Z-value: 0.44
Transcription factors associated with ZNF350
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF350
|
ENSG00000256683.2 | ZNF350 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF350 | hg19_v2_chr19_-_52489923_52490113 | -0.50 | 4.7e-02 | Click! |
Activity profile of ZNF350 motif
Sorted Z-values of ZNF350 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF350
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_39674668 | 1.45 |
ENST00000393981.3 |
KRT15 |
keratin 15 |
| chr11_-_112034803 | 0.74 |
ENST00000528832.1 |
IL18 |
interleukin 18 (interferon-gamma-inducing factor) |
| chr11_-_112034831 | 0.70 |
ENST00000280357.7 |
IL18 |
interleukin 18 (interferon-gamma-inducing factor) |
| chr11_-_112034780 | 0.69 |
ENST00000524595.1 |
IL18 |
interleukin 18 (interferon-gamma-inducing factor) |
| chr2_+_235860616 | 0.59 |
ENST00000392011.2 |
SH3BP4 |
SH3-domain binding protein 4 |
| chr20_+_4666882 | 0.54 |
ENST00000379440.4 ENST00000430350.2 |
PRNP |
prion protein |
| chr2_-_219157250 | 0.46 |
ENST00000434015.2 ENST00000444183.1 ENST00000420341.1 ENST00000453281.1 ENST00000258412.3 ENST00000440422.1 |
TMBIM1 |
transmembrane BAX inhibitor motif containing 1 |
| chr20_+_4667094 | 0.41 |
ENST00000424424.1 ENST00000457586.1 |
PRNP |
prion protein |
| chr20_+_44035847 | 0.41 |
ENST00000372712.2 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr4_-_99578789 | 0.40 |
ENST00000511651.1 ENST00000505184.1 |
TSPAN5 |
tetraspanin 5 |
| chr16_+_4526341 | 0.35 |
ENST00000458134.3 ENST00000219700.6 ENST00000575120.1 ENST00000572812.1 ENST00000574466.1 ENST00000576827.1 ENST00000570445.1 |
HMOX2 |
heme oxygenase (decycling) 2 |
| chr12_+_113376157 | 0.35 |
ENST00000228928.7 |
OAS3 |
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr4_-_99578776 | 0.33 |
ENST00000515287.1 |
TSPAN5 |
tetraspanin 5 |
| chr20_+_44035200 | 0.33 |
ENST00000372717.1 ENST00000360981.4 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr17_-_62340581 | 0.32 |
ENST00000258991.3 ENST00000583738.1 ENST00000584379.1 |
TEX2 |
testis expressed 2 |
| chr3_-_155524049 | 0.30 |
ENST00000534941.1 ENST00000340171.2 |
C3orf33 |
chromosome 3 open reading frame 33 |
| chr11_-_327537 | 0.30 |
ENST00000602735.1 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr4_-_120133661 | 0.27 |
ENST00000503243.1 ENST00000326780.3 |
RP11-455G16.1 |
Uncharacterized protein |
| chr2_-_9143786 | 0.24 |
ENST00000462696.1 ENST00000305997.3 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
| chr14_+_96671016 | 0.21 |
ENST00000542454.2 ENST00000554311.1 ENST00000306005.3 ENST00000539359.1 ENST00000553811.1 |
BDKRB2 RP11-404P21.8 |
bradykinin receptor B2 Uncharacterized protein |
| chr2_-_21266935 | 0.21 |
ENST00000233242.1 |
APOB |
apolipoprotein B |
| chr17_+_48624450 | 0.21 |
ENST00000006658.6 ENST00000356488.4 ENST00000393244.3 |
SPATA20 |
spermatogenesis associated 20 |
| chr19_-_5680499 | 0.16 |
ENST00000587589.1 |
C19orf70 |
chromosome 19 open reading frame 70 |
| chr8_-_119124045 | 0.13 |
ENST00000378204.2 |
EXT1 |
exostosin glycosyltransferase 1 |
| chr19_+_35645817 | 0.11 |
ENST00000423817.3 |
FXYD5 |
FXYD domain containing ion transport regulator 5 |
| chr16_+_14802801 | 0.11 |
ENST00000526520.1 ENST00000531598.2 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr19_+_35645618 | 0.11 |
ENST00000392218.2 ENST00000543307.1 ENST00000392219.2 ENST00000541435.2 ENST00000590686.1 ENST00000342879.3 ENST00000588699.1 |
FXYD5 |
FXYD domain containing ion transport regulator 5 |
| chr12_+_81471816 | 0.11 |
ENST00000261206.3 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
| chr20_-_35374456 | 0.11 |
ENST00000373803.2 ENST00000359675.2 ENST00000540765.1 ENST00000349004.1 |
NDRG3 |
NDRG family member 3 |
| chr2_+_219524379 | 0.10 |
ENST00000443791.1 ENST00000359273.3 ENST00000392109.1 ENST00000392110.2 ENST00000423377.1 ENST00000392111.2 ENST00000412366.1 |
BCS1L |
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr22_-_50964558 | 0.09 |
ENST00000535425.1 ENST00000439934.1 |
SCO2 |
SCO2 cytochrome c oxidase assembly protein |
| chr11_-_7041466 | 0.09 |
ENST00000536068.1 ENST00000278314.4 |
ZNF214 |
zinc finger protein 214 |
| chr21_-_30365136 | 0.09 |
ENST00000361371.5 ENST00000389194.2 ENST00000389195.2 |
LTN1 |
listerin E3 ubiquitin protein ligase 1 |
| chr19_+_2977444 | 0.08 |
ENST00000246112.4 ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6 |
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr11_-_69633792 | 0.08 |
ENST00000334134.2 |
FGF3 |
fibroblast growth factor 3 |
| chr3_-_128902729 | 0.08 |
ENST00000451728.2 ENST00000446936.2 ENST00000502976.1 ENST00000500450.2 ENST00000441626.2 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
| chr15_+_42694573 | 0.07 |
ENST00000397200.4 ENST00000569827.1 |
CAPN3 |
calpain 3, (p94) |
| chr1_-_200379180 | 0.06 |
ENST00000294740.3 |
ZNF281 |
zinc finger protein 281 |
| chr11_-_8954491 | 0.06 |
ENST00000526227.1 ENST00000525780.1 ENST00000326053.5 |
C11orf16 |
chromosome 11 open reading frame 16 |
| chr19_+_1450112 | 0.06 |
ENST00000590469.1 ENST00000233607.2 ENST00000238483.4 ENST00000590877.1 |
APC2 |
adenomatosis polyposis coli 2 |
| chr17_-_18950310 | 0.06 |
ENST00000573099.1 |
GRAP |
GRB2-related adaptor protein |
| chr2_-_106810783 | 0.06 |
ENST00000283148.7 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
| chr19_-_49243845 | 0.06 |
ENST00000222145.4 |
RASIP1 |
Ras interacting protein 1 |
| chr2_+_219524473 | 0.06 |
ENST00000439945.1 ENST00000431802.1 |
BCS1L |
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr17_-_6554877 | 0.05 |
ENST00000225728.3 ENST00000575197.1 |
MED31 |
mediator complex subunit 31 |
| chr10_+_50822480 | 0.05 |
ENST00000455728.2 |
CHAT |
choline O-acetyltransferase |
| chr2_-_70780770 | 0.05 |
ENST00000444975.1 ENST00000445399.1 ENST00000418333.2 |
TGFA |
transforming growth factor, alpha |
| chr2_-_106810742 | 0.05 |
ENST00000409501.3 ENST00000428048.2 ENST00000441952.1 ENST00000457835.1 ENST00000540130.1 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
| chr8_-_144679296 | 0.05 |
ENST00000317198.6 |
EEF1D |
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr11_+_68671310 | 0.05 |
ENST00000255078.3 ENST00000539224.1 |
IGHMBP2 |
immunoglobulin mu binding protein 2 |
| chr1_-_33647267 | 0.05 |
ENST00000291416.5 |
TRIM62 |
tripartite motif containing 62 |
| chrX_-_47479246 | 0.04 |
ENST00000295987.7 ENST00000340666.4 |
SYN1 |
synapsin I |
| chr1_+_204797749 | 0.04 |
ENST00000367172.4 ENST00000367171.4 ENST00000367170.4 ENST00000338515.6 ENST00000339876.6 ENST00000338586.6 ENST00000539706.1 ENST00000360049.4 ENST00000367169.4 ENST00000446412.1 ENST00000403080.1 |
NFASC |
neurofascin |
| chr11_+_8932828 | 0.04 |
ENST00000530281.1 ENST00000396648.2 ENST00000534147.1 ENST00000529942.1 |
AKIP1 |
A kinase (PRKA) interacting protein 1 |
| chr17_-_39597173 | 0.04 |
ENST00000246646.3 |
KRT38 |
keratin 38 |
| chr2_-_70780572 | 0.03 |
ENST00000450929.1 |
TGFA |
transforming growth factor, alpha |
| chr1_+_156611960 | 0.03 |
ENST00000361588.5 |
BCAN |
brevican |
| chrX_+_120181457 | 0.03 |
ENST00000328078.1 |
GLUD2 |
glutamate dehydrogenase 2 |
| chr1_-_200379129 | 0.03 |
ENST00000367353.1 |
ZNF281 |
zinc finger protein 281 |
| chr11_+_64323428 | 0.03 |
ENST00000377581.3 |
SLC22A11 |
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr12_-_93323013 | 0.03 |
ENST00000322349.8 |
EEA1 |
early endosome antigen 1 |
| chr16_+_31225337 | 0.03 |
ENST00000322122.3 |
TRIM72 |
tripartite motif containing 72 |
| chr3_-_128902759 | 0.03 |
ENST00000422453.2 ENST00000504813.1 ENST00000512338.1 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
| chr6_-_28303901 | 0.03 |
ENST00000439158.1 ENST00000446474.1 ENST00000414431.1 ENST00000344279.6 ENST00000453745.1 |
ZSCAN31 |
zinc finger and SCAN domain containing 31 |
| chr1_-_247094628 | 0.03 |
ENST00000366508.1 ENST00000326225.3 ENST00000391829.2 |
AHCTF1 |
AT hook containing transcription factor 1 |
| chr15_-_43029252 | 0.03 |
ENST00000563260.1 ENST00000356231.3 |
CDAN1 |
codanin 1 |
| chr1_+_63788730 | 0.03 |
ENST00000371116.2 |
FOXD3 |
forkhead box D3 |
| chr17_-_18950950 | 0.03 |
ENST00000284154.5 |
GRAP |
GRB2-related adaptor protein |
| chr10_-_15762124 | 0.03 |
ENST00000378076.3 |
ITGA8 |
integrin, alpha 8 |
| chr11_+_8932715 | 0.03 |
ENST00000529876.1 ENST00000525005.1 ENST00000524577.1 ENST00000534506.1 |
AKIP1 |
A kinase (PRKA) interacting protein 1 |
| chr2_+_238600788 | 0.02 |
ENST00000289175.6 ENST00000244815.5 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
| chr17_-_8093471 | 0.02 |
ENST00000389017.4 |
C17orf59 |
chromosome 17 open reading frame 59 |
| chr1_+_156611704 | 0.02 |
ENST00000329117.5 |
BCAN |
brevican |
| chr14_+_73603126 | 0.02 |
ENST00000557356.1 ENST00000556864.1 ENST00000556533.1 ENST00000556951.1 ENST00000557293.1 ENST00000553719.1 ENST00000553599.1 ENST00000556011.1 ENST00000394157.3 ENST00000357710.4 ENST00000324501.5 ENST00000560005.2 ENST00000555254.1 ENST00000261970.3 ENST00000344094.3 ENST00000554131.1 ENST00000557037.1 |
PSEN1 |
presenilin 1 |
| chr10_+_50822083 | 0.02 |
ENST00000337653.2 ENST00000395562.2 |
CHAT |
choline O-acetyltransferase |
| chr16_-_46865047 | 0.01 |
ENST00000394806.2 |
C16orf87 |
chromosome 16 open reading frame 87 |
| chr3_-_11623804 | 0.01 |
ENST00000451674.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr19_+_5681011 | 0.01 |
ENST00000581893.1 ENST00000411793.2 ENST00000301382.4 ENST00000581773.1 ENST00000423665.2 ENST00000583928.1 ENST00000342970.2 ENST00000422535.2 ENST00000581521.1 ENST00000339423.2 |
HSD11B1L |
hydroxysteroid (11-beta) dehydrogenase 1-like |
| chr11_-_64647144 | 0.01 |
ENST00000359393.2 ENST00000433803.1 ENST00000411683.1 |
EHD1 |
EH-domain containing 1 |
| chr2_+_177025619 | 0.01 |
ENST00000410016.1 |
HOXD3 |
homeobox D3 |
| chr10_-_12084770 | 0.01 |
ENST00000357604.5 |
UPF2 |
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr7_+_2559399 | 0.01 |
ENST00000222725.5 ENST00000359574.3 |
LFNG |
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr19_-_4535233 | 0.01 |
ENST00000381848.3 ENST00000588887.1 ENST00000586133.1 |
PLIN5 |
perilipin 5 |
| chr10_-_120938303 | 0.00 |
ENST00000356951.3 ENST00000298510.2 |
PRDX3 |
peroxiredoxin 3 |
| chr19_+_49866851 | 0.00 |
ENST00000221498.2 ENST00000596402.1 |
DKKL1 |
dickkopf-like 1 |
| chr1_-_204654481 | 0.00 |
ENST00000367176.3 |
LRRN2 |
leucine rich repeat neuronal 2 |
| chr16_+_67282853 | 0.00 |
ENST00000299798.11 |
SLC9A5 |
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.3 | 1.0 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 0.5 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.2 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.3 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.2 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.6 | GO:0043090 | amino acid import(GO:0043090) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 1.5 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.0 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.1 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0004102 | choline O-acetyltransferase activity(GO:0004102) |
| 0.0 | 1.5 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.5 | GO:0005123 | death receptor binding(GO:0005123) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 2.1 | REACTOME SIGNALING BY ILS | Genes involved in Signaling by Interleukins |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |