Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
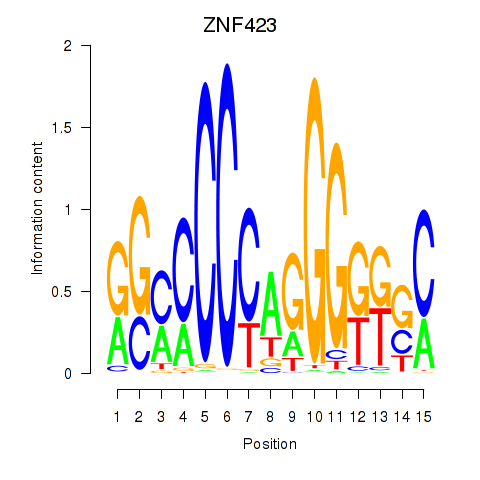
Results for ZNF423
Z-value: 0.85
Transcription factors associated with ZNF423
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF423
|
ENSG00000102935.7 | ZNF423 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF423 | hg19_v2_chr16_-_49698136_49698209 | 0.27 | 3.0e-01 | Click! |
Activity profile of ZNF423 motif
Sorted Z-values of ZNF423 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF423
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_135385202 | 2.03 |
ENST00000514554.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr10_+_17271266 | 1.97 |
ENST00000224237.5 |
VIM |
vimentin |
| chr2_+_46769798 | 1.62 |
ENST00000238738.4 |
RHOQ |
ras homolog family member Q |
| chrX_+_49028265 | 1.49 |
ENST00000376322.3 ENST00000376327.5 |
PLP2 |
proteolipid protein 2 (colonic epithelium-enriched) |
| chrX_-_51239425 | 1.46 |
ENST00000375992.3 |
NUDT11 |
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr9_+_112542591 | 1.35 |
ENST00000483909.1 ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2 PALM2-AKAP2 AKAP2 |
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr9_+_112542572 | 1.28 |
ENST00000374530.3 |
PALM2-AKAP2 |
PALM2-AKAP2 readthrough |
| chr10_-_121296045 | 1.13 |
ENST00000392865.1 |
RGS10 |
regulator of G-protein signaling 10 |
| chr3_-_69129501 | 1.05 |
ENST00000540295.1 ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3 |
ubiquitin-like modifier activating enzyme 3 |
| chr11_-_44972390 | 0.78 |
ENST00000395648.3 ENST00000531928.2 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr11_-_61658853 | 0.77 |
ENST00000525588.1 ENST00000540820.1 |
FADS3 |
fatty acid desaturase 3 |
| chr11_-_61659006 | 0.64 |
ENST00000278829.2 |
FADS3 |
fatty acid desaturase 3 |
| chr9_-_16870704 | 0.64 |
ENST00000380672.4 ENST00000380667.2 ENST00000380666.2 ENST00000486514.1 |
BNC2 |
basonuclin 2 |
| chrX_-_107019181 | 0.61 |
ENST00000315660.4 ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr10_-_75255724 | 0.60 |
ENST00000342558.3 ENST00000360663.5 ENST00000394829.2 ENST00000394828.2 ENST00000394822.2 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chrX_-_107018969 | 0.55 |
ENST00000372383.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr1_-_203055129 | 0.54 |
ENST00000241651.4 |
MYOG |
myogenin (myogenic factor 4) |
| chr16_-_11681316 | 0.54 |
ENST00000571688.1 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr11_-_44972476 | 0.53 |
ENST00000527685.1 ENST00000308212.5 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr19_-_15311713 | 0.53 |
ENST00000601011.1 ENST00000263388.2 |
NOTCH3 |
notch 3 |
| chr7_-_30029367 | 0.50 |
ENST00000242059.5 |
SCRN1 |
secernin 1 |
| chr11_-_67120974 | 0.47 |
ENST00000539074.1 ENST00000312419.3 |
POLD4 |
polymerase (DNA-directed), delta 4, accessory subunit |
| chr11_-_44971702 | 0.46 |
ENST00000533940.1 ENST00000533937.1 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chrX_-_1572629 | 0.45 |
ENST00000534940.1 |
ASMTL |
acetylserotonin O-methyltransferase-like |
| chr8_+_22022223 | 0.43 |
ENST00000306385.5 |
BMP1 |
bone morphogenetic protein 1 |
| chr7_-_5570229 | 0.42 |
ENST00000331789.5 |
ACTB |
actin, beta |
| chr11_+_73019282 | 0.40 |
ENST00000263674.3 |
ARHGEF17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr16_-_11681023 | 0.39 |
ENST00000570904.1 ENST00000574701.1 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr8_-_103668114 | 0.38 |
ENST00000285407.6 |
KLF10 |
Kruppel-like factor 10 |
| chr10_-_118032979 | 0.37 |
ENST00000355422.6 |
GFRA1 |
GDNF family receptor alpha 1 |
| chr5_-_172756506 | 0.36 |
ENST00000265087.4 |
STC2 |
stanniocalcin 2 |
| chr21_-_18985230 | 0.36 |
ENST00000457956.1 ENST00000348354.6 |
BTG3 |
BTG family, member 3 |
| chr1_-_155177677 | 0.35 |
ENST00000368378.3 ENST00000541990.1 ENST00000457183.2 |
THBS3 |
thrombospondin 3 |
| chr10_-_75255668 | 0.34 |
ENST00000545874.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr12_-_57522813 | 0.33 |
ENST00000556155.1 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr3_-_52931557 | 0.33 |
ENST00000504329.1 ENST00000355083.5 |
TMEM110-MUSTN1 TMEM110 |
TMEM110-MUSTN1 readthrough transmembrane protein 110 |
| chr15_+_100106244 | 0.32 |
ENST00000557942.1 |
MEF2A |
myocyte enhancer factor 2A |
| chr19_+_41770236 | 0.31 |
ENST00000392006.3 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr15_+_100106155 | 0.30 |
ENST00000557785.1 ENST00000558049.1 ENST00000449277.2 |
MEF2A |
myocyte enhancer factor 2A |
| chr19_-_11689752 | 0.27 |
ENST00000592659.1 ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr6_-_91296602 | 0.27 |
ENST00000369325.3 ENST00000369327.3 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
| chr9_-_99382065 | 0.26 |
ENST00000265659.2 ENST00000375241.1 ENST00000375236.1 |
CDC14B |
cell division cycle 14B |
| chr6_-_91296737 | 0.26 |
ENST00000369332.3 ENST00000369329.3 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
| chr9_-_99381660 | 0.25 |
ENST00000375240.3 ENST00000463569.1 |
CDC14B |
cell division cycle 14B |
| chr12_-_118810688 | 0.25 |
ENST00000542532.1 ENST00000392533.3 |
TAOK3 |
TAO kinase 3 |
| chr22_-_43045574 | 0.24 |
ENST00000352397.5 |
CYB5R3 |
cytochrome b5 reductase 3 |
| chr8_+_22423168 | 0.24 |
ENST00000518912.1 ENST00000428103.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr21_-_18985158 | 0.23 |
ENST00000339775.6 |
BTG3 |
BTG family, member 3 |
| chr6_-_32160622 | 0.23 |
ENST00000487761.1 ENST00000375040.3 |
GPSM3 |
G-protein signaling modulator 3 |
| chr15_-_59225758 | 0.22 |
ENST00000558486.1 ENST00000560682.1 ENST00000249736.7 ENST00000559880.1 ENST00000536328.1 |
SLTM |
SAFB-like, transcription modulator |
| chr2_-_120980939 | 0.22 |
ENST00000426077.2 |
TMEM185B |
transmembrane protein 185B |
| chr11_-_64013663 | 0.22 |
ENST00000392210.2 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr22_+_35653445 | 0.21 |
ENST00000420166.1 ENST00000444518.2 ENST00000455359.1 ENST00000216106.5 |
HMGXB4 |
HMG box domain containing 4 |
| chr6_-_143266297 | 0.19 |
ENST00000367603.2 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
| chr11_-_6495082 | 0.19 |
ENST00000536344.1 |
TRIM3 |
tripartite motif containing 3 |
| chr11_-_64510409 | 0.19 |
ENST00000394429.1 ENST00000394428.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr10_+_135340859 | 0.18 |
ENST00000252945.3 ENST00000421586.1 ENST00000418356.1 |
CYP2E1 |
cytochrome P450, family 2, subfamily E, polypeptide 1 |
| chr19_-_57183114 | 0.18 |
ENST00000537055.2 ENST00000601659.1 |
ZNF835 |
zinc finger protein 835 |
| chr10_+_88428206 | 0.16 |
ENST00000429277.2 ENST00000458213.2 ENST00000352360.5 |
LDB3 |
LIM domain binding 3 |
| chr4_+_4388805 | 0.16 |
ENST00000504171.1 |
NSG1 |
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr19_-_10679644 | 0.15 |
ENST00000393599.2 |
CDKN2D |
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr17_+_6347729 | 0.15 |
ENST00000572447.1 |
FAM64A |
family with sequence similarity 64, member A |
| chr15_+_41523335 | 0.14 |
ENST00000334660.5 |
CHP1 |
calcineurin-like EF-hand protein 1 |
| chr4_+_4387983 | 0.14 |
ENST00000397958.1 |
NSG1 |
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chr17_+_77020325 | 0.14 |
ENST00000311661.4 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
| chr15_-_59225844 | 0.12 |
ENST00000380516.2 |
SLTM |
SAFB-like, transcription modulator |
| chr2_+_62932779 | 0.12 |
ENST00000427809.1 ENST00000405482.1 ENST00000431489.1 |
EHBP1 |
EH domain binding protein 1 |
| chr11_-_1036706 | 0.12 |
ENST00000421673.2 |
MUC6 |
mucin 6, oligomeric mucus/gel-forming |
| chr8_+_22422749 | 0.12 |
ENST00000523900.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr17_+_77020224 | 0.11 |
ENST00000339142.2 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
| chr17_+_6347761 | 0.11 |
ENST00000250056.8 ENST00000571373.1 ENST00000570337.2 ENST00000572595.2 ENST00000576056.1 |
FAM64A |
family with sequence similarity 64, member A |
| chr19_+_41770269 | 0.11 |
ENST00000378215.4 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr4_+_4388245 | 0.10 |
ENST00000433139.2 |
NSG1 |
Homo sapiens neuron specific gene family member 1 (NSG1), transcript variant 3, mRNA. |
| chrX_+_136648297 | 0.10 |
ENST00000287538.5 |
ZIC3 |
Zic family member 3 |
| chr8_+_22022653 | 0.09 |
ENST00000354870.5 ENST00000397816.3 ENST00000306349.8 |
BMP1 |
bone morphogenetic protein 1 |
| chr11_-_6495101 | 0.09 |
ENST00000528227.1 ENST00000359518.3 ENST00000345851.3 ENST00000537602.1 |
TRIM3 |
tripartite motif containing 3 |
| chr5_+_141303373 | 0.09 |
ENST00000432126.2 ENST00000194118.4 |
KIAA0141 |
KIAA0141 |
| chr11_-_44972418 | 0.08 |
ENST00000525680.1 ENST00000528290.1 ENST00000530035.1 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr8_+_30241995 | 0.08 |
ENST00000397323.4 ENST00000339877.4 ENST00000320203.4 ENST00000287771.5 |
RBPMS |
RNA binding protein with multiple splicing |
| chr17_-_73179046 | 0.07 |
ENST00000314523.7 ENST00000420826.2 |
SUMO2 |
small ubiquitin-like modifier 2 |
| chr1_+_1222489 | 0.07 |
ENST00000379099.3 |
SCNN1D |
sodium channel, non-voltage-gated 1, delta subunit |
| chr3_-_48594248 | 0.07 |
ENST00000545984.1 ENST00000232375.3 ENST00000416568.1 ENST00000383734.2 ENST00000541519.1 ENST00000412035.1 |
PFKFB4 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr6_-_35480705 | 0.06 |
ENST00000229771.6 |
TULP1 |
tubby like protein 1 |
| chr3_+_32726774 | 0.06 |
ENST00000538368.1 |
CNOT10 |
CCR4-NOT transcription complex, subunit 10 |
| chr3_+_180630444 | 0.06 |
ENST00000491062.1 ENST00000468861.1 ENST00000445140.2 ENST00000484958.1 |
FXR1 |
fragile X mental retardation, autosomal homolog 1 |
| chr16_+_333152 | 0.06 |
ENST00000219406.6 ENST00000404312.1 ENST00000456379.1 |
PDIA2 |
protein disulfide isomerase family A, member 2 |
| chr8_+_30241934 | 0.06 |
ENST00000538486.1 |
RBPMS |
RNA binding protein with multiple splicing |
| chr17_-_73178599 | 0.06 |
ENST00000578238.1 |
SUMO2 |
small ubiquitin-like modifier 2 |
| chr15_-_83316711 | 0.05 |
ENST00000568128.1 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr20_+_43029911 | 0.05 |
ENST00000443598.2 ENST00000316099.4 ENST00000415691.2 |
HNF4A |
hepatocyte nuclear factor 4, alpha |
| chr1_+_155278625 | 0.05 |
ENST00000368356.4 ENST00000356657.6 |
FDPS |
farnesyl diphosphate synthase |
| chr16_+_27325202 | 0.04 |
ENST00000395762.2 ENST00000562142.1 ENST00000561742.1 ENST00000543915.2 ENST00000449195.1 ENST00000380922.3 ENST00000563002.1 |
IL4R |
interleukin 4 receptor |
| chr10_+_124913930 | 0.03 |
ENST00000368858.5 |
BUB3 |
BUB3 mitotic checkpoint protein |
| chr15_-_66649010 | 0.03 |
ENST00000367709.4 ENST00000261881.4 |
TIPIN |
TIMELESS interacting protein |
| chr19_+_10531150 | 0.03 |
ENST00000352831.6 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr10_-_76859247 | 0.03 |
ENST00000472493.2 ENST00000605915.1 ENST00000478873.2 |
DUSP13 |
dual specificity phosphatase 13 |
| chr19_+_41770349 | 0.03 |
ENST00000602130.1 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr1_+_10459111 | 0.03 |
ENST00000541529.1 ENST00000270776.8 ENST00000483936.1 ENST00000538557.1 |
PGD |
phosphogluconate dehydrogenase |
| chr19_-_23941639 | 0.02 |
ENST00000395385.3 ENST00000531570.1 ENST00000528059.1 |
ZNF681 |
zinc finger protein 681 |
| chr15_+_41523417 | 0.02 |
ENST00000560397.1 |
CHP1 |
calcineurin-like EF-hand protein 1 |
| chr7_-_100491854 | 0.02 |
ENST00000426415.1 ENST00000430554.1 ENST00000412389.1 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr7_-_100253993 | 0.01 |
ENST00000461605.1 ENST00000160382.5 |
ACTL6B |
actin-like 6B |
| chr7_-_27219849 | 0.01 |
ENST00000396344.4 |
HOXA10 |
homeobox A10 |
| chr8_+_22423219 | 0.01 |
ENST00000523965.1 ENST00000521554.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr19_+_20011775 | 0.00 |
ENST00000592245.1 ENST00000592160.1 ENST00000343769.5 |
AC007204.2 ZNF93 |
AC007204.2 zinc finger protein 93 |
| chr19_-_23941680 | 0.00 |
ENST00000402377.3 |
ZNF681 |
zinc finger protein 681 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 2.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 2.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 0.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.9 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 0.4 | GO:0098845 | postsynaptic endosome(GO:0098845) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.4 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 1.6 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 2.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.9 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.3 | 2.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 1.5 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.1 | 1.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 1.2 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.9 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.4 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 1.5 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 1.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.4 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 1.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.5 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.0 | 0.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0008391 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.3 | 1.5 | GO:1901911 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 2.0 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.2 | 0.5 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 1.2 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.6 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 1.6 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 0.3 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.9 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.5 | GO:0072104 | glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 1.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.1 | 0.3 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.4 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) |
| 0.0 | 0.5 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.4 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 1.0 | GO:0003416 | endochondral bone growth(GO:0003416) |
| 0.0 | 0.9 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.5 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.5 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.0 | 0.2 | GO:0070885 | positive regulation of sodium:proton antiporter activity(GO:0032417) negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 1.4 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 2.0 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.1 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.5 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 0.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.0 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) response to odorant(GO:1990834) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 2.6 | GO:0008360 | regulation of cell shape(GO:0008360) |