Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
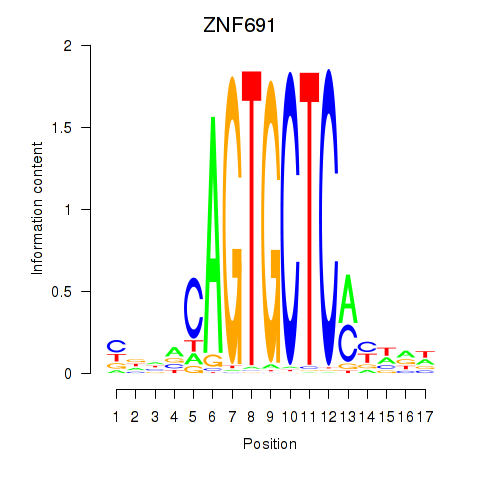
Results for ZNF691
Z-value: 0.44
Transcription factors associated with ZNF691
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF691
|
ENSG00000164011.13 | ZNF691 |
Activity profile of ZNF691 motif
Sorted Z-values of ZNF691 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF691
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_23264766 | 1.58 |
ENST00000390331.2 |
IGLC7 |
immunoglobulin lambda constant 7 |
| chr11_+_60223312 | 1.06 |
ENST00000532491.1 ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr11_+_60223225 | 1.05 |
ENST00000524807.1 ENST00000345732.4 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr1_+_26644441 | 0.96 |
ENST00000374213.2 |
CD52 |
CD52 molecule |
| chr10_-_91011548 | 0.72 |
ENST00000456827.1 |
LIPA |
lipase A, lysosomal acid, cholesterol esterase |
| chr11_+_102188272 | 0.70 |
ENST00000532808.1 |
BIRC3 |
baculoviral IAP repeat containing 3 |
| chr11_+_121461097 | 0.60 |
ENST00000527934.1 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr1_+_28199047 | 0.59 |
ENST00000373925.1 ENST00000328928.7 ENST00000373927.3 ENST00000427466.1 ENST00000442118.1 ENST00000373921.3 |
THEMIS2 |
thymocyte selection associated family member 2 |
| chr12_-_51717875 | 0.55 |
ENST00000604560.1 |
BIN2 |
bridging integrator 2 |
| chr2_-_110371412 | 0.54 |
ENST00000415095.1 ENST00000334001.6 ENST00000437928.1 ENST00000493445.1 ENST00000397714.2 ENST00000461295.1 |
SEPT10 |
septin 10 |
| chr12_-_51717948 | 0.46 |
ENST00000267012.4 |
BIN2 |
bridging integrator 2 |
| chr12_-_51717922 | 0.45 |
ENST00000452142.2 |
BIN2 |
bridging integrator 2 |
| chr1_+_26856236 | 0.44 |
ENST00000374168.2 ENST00000374166.4 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr14_-_106725723 | 0.42 |
ENST00000390609.2 |
IGHV3-23 |
immunoglobulin heavy variable 3-23 |
| chr8_+_27168988 | 0.41 |
ENST00000397501.1 ENST00000338238.4 ENST00000544172.1 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr2_-_110371720 | 0.40 |
ENST00000356688.4 |
SEPT10 |
septin 10 |
| chr2_-_110371664 | 0.39 |
ENST00000545389.1 ENST00000423520.1 |
SEPT10 |
septin 10 |
| chr8_-_87526561 | 0.38 |
ENST00000523911.1 |
RMDN1 |
regulator of microtubule dynamics 1 |
| chr4_-_120243545 | 0.35 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr19_-_51472222 | 0.28 |
ENST00000376851.3 |
KLK6 |
kallikrein-related peptidase 6 |
| chr17_-_61777459 | 0.28 |
ENST00000578993.1 ENST00000583211.1 ENST00000259006.3 |
LIMD2 |
LIM domain containing 2 |
| chr17_-_41738931 | 0.27 |
ENST00000329168.3 ENST00000549132.1 |
MEOX1 |
mesenchyme homeobox 1 |
| chr6_-_159240415 | 0.25 |
ENST00000367075.3 |
EZR |
ezrin |
| chr1_+_203734296 | 0.21 |
ENST00000442561.2 ENST00000367217.5 |
LAX1 |
lymphocyte transmembrane adaptor 1 |
| chr3_+_44596679 | 0.20 |
ENST00000426540.1 ENST00000431636.1 ENST00000341840.3 ENST00000273320.3 |
ZKSCAN7 |
zinc finger with KRAB and SCAN domains 7 |
| chr1_+_15986364 | 0.17 |
ENST00000345034.1 |
RSC1A1 |
regulatory solute carrier protein, family 1, member 1 |
| chr3_-_167452298 | 0.17 |
ENST00000475915.2 ENST00000462725.2 ENST00000461494.1 |
PDCD10 |
programmed cell death 10 |
| chr9_+_135754263 | 0.17 |
ENST00000356311.5 ENST00000350499.6 |
C9orf9 |
chromosome 9 open reading frame 9 |
| chr6_-_144329531 | 0.16 |
ENST00000429150.1 ENST00000392309.1 ENST00000416623.1 ENST00000392307.1 |
PLAGL1 |
pleiomorphic adenoma gene-like 1 |
| chr3_-_167452614 | 0.16 |
ENST00000392750.2 ENST00000464360.1 ENST00000492139.1 ENST00000471885.1 ENST00000470131.1 |
PDCD10 |
programmed cell death 10 |
| chr19_-_51472031 | 0.16 |
ENST00000391808.1 |
KLK6 |
kallikrein-related peptidase 6 |
| chr1_+_145516560 | 0.15 |
ENST00000537888.1 |
PEX11B |
peroxisomal biogenesis factor 11 beta |
| chr19_+_50380917 | 0.15 |
ENST00000535102.2 |
TBC1D17 |
TBC1 domain family, member 17 |
| chr12_-_51718436 | 0.14 |
ENST00000544402.1 |
BIN2 |
bridging integrator 2 |
| chr1_-_203144941 | 0.12 |
ENST00000255416.4 |
MYBPH |
myosin binding protein H |
| chr3_-_167452262 | 0.12 |
ENST00000487947.2 |
PDCD10 |
programmed cell death 10 |
| chr12_+_120502441 | 0.12 |
ENST00000446727.2 |
CCDC64 |
coiled-coil domain containing 64 |
| chr11_-_133826852 | 0.12 |
ENST00000533871.2 ENST00000321016.8 |
IGSF9B |
immunoglobulin superfamily, member 9B |
| chr15_-_74726283 | 0.11 |
ENST00000543145.2 |
SEMA7A |
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr1_+_154966058 | 0.11 |
ENST00000392487.1 |
LENEP |
lens epithelial protein |
| chr19_-_17932314 | 0.10 |
ENST00000598577.1 ENST00000317306.7 ENST00000379695.5 |
INSL3 |
insulin-like 3 (Leydig cell) |
| chr19_-_50311896 | 0.10 |
ENST00000529634.2 |
FUZ |
fuzzy planar cell polarity protein |
| chr16_-_1821496 | 0.09 |
ENST00000564628.1 ENST00000563498.1 |
NME3 |
NME/NM23 nucleoside diphosphate kinase 3 |
| chr19_-_5903714 | 0.09 |
ENST00000586349.1 ENST00000585661.1 ENST00000308961.4 ENST00000592634.1 ENST00000418389.2 ENST00000252675.5 |
AC024592.12 NDUFA11 FUT5 |
Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa fucosyltransferase 5 (alpha (1,3) fucosyltransferase) |
| chr11_-_65655906 | 0.08 |
ENST00000533045.1 ENST00000338369.2 ENST00000357519.4 |
FIBP |
fibroblast growth factor (acidic) intracellular binding protein |
| chr13_-_52027134 | 0.08 |
ENST00000311234.4 ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6 |
integrator complex subunit 6 |
| chr19_+_50084561 | 0.08 |
ENST00000246794.5 |
PRRG2 |
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr8_+_32579341 | 0.07 |
ENST00000519240.1 ENST00000539990.1 |
NRG1 |
neuregulin 1 |
| chr7_-_143059780 | 0.07 |
ENST00000409578.1 ENST00000409346.1 |
FAM131B |
family with sequence similarity 131, member B |
| chr2_+_238475217 | 0.06 |
ENST00000165524.1 |
PRLH |
prolactin releasing hormone |
| chr9_+_37486005 | 0.06 |
ENST00000377792.3 |
POLR1E |
polymerase (RNA) I polypeptide E, 53kDa |
| chr17_+_30771279 | 0.06 |
ENST00000261712.3 ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr9_+_37485932 | 0.06 |
ENST00000377798.4 ENST00000442009.2 |
POLR1E |
polymerase (RNA) I polypeptide E, 53kDa |
| chr4_+_657485 | 0.05 |
ENST00000471824.2 |
PDE6B |
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr1_-_203055129 | 0.05 |
ENST00000241651.4 |
MYOG |
myogenin (myogenic factor 4) |
| chr8_-_144679532 | 0.05 |
ENST00000534380.1 ENST00000533494.1 ENST00000531218.1 ENST00000526340.1 ENST00000533204.1 ENST00000532400.1 ENST00000529516.1 ENST00000534377.1 ENST00000531621.1 ENST00000530191.1 ENST00000524900.1 ENST00000526838.1 ENST00000531931.1 ENST00000534475.1 ENST00000442189.2 ENST00000524624.1 ENST00000532596.1 ENST00000529832.1 ENST00000530306.1 ENST00000530545.1 ENST00000525261.1 ENST00000534804.1 ENST00000528303.1 ENST00000528610.1 |
EEF1D |
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr3_+_129693523 | 0.05 |
ENST00000507066.1 |
TRH |
thyrotropin-releasing hormone |
| chr11_-_36619771 | 0.05 |
ENST00000311485.3 ENST00000527033.1 ENST00000532616.1 |
RAG2 |
recombination activating gene 2 |
| chr21_-_37852359 | 0.05 |
ENST00000399137.1 ENST00000399135.1 |
CLDN14 |
claudin 14 |
| chr6_+_31553978 | 0.05 |
ENST00000376096.1 ENST00000376099.1 ENST00000376110.3 |
LST1 |
leukocyte specific transcript 1 |
| chr20_-_1317555 | 0.05 |
ENST00000537552.1 |
AL136531.1 |
HCG2043693; Uncharacterized protein |
| chr5_-_131347501 | 0.04 |
ENST00000543479.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr2_-_203736334 | 0.04 |
ENST00000392237.2 ENST00000416760.1 ENST00000412210.1 |
ICA1L |
islet cell autoantigen 1,69kDa-like |
| chr2_+_68961905 | 0.04 |
ENST00000295381.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr11_+_64949899 | 0.04 |
ENST00000531068.1 ENST00000527699.1 ENST00000533909.1 ENST00000527323.1 |
CAPN1 |
calpain 1, (mu/I) large subunit |
| chr6_+_31553901 | 0.04 |
ENST00000418507.2 ENST00000438075.2 ENST00000376100.3 ENST00000376111.4 |
LST1 |
leukocyte specific transcript 1 |
| chr5_-_179045199 | 0.04 |
ENST00000523921.1 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr12_-_12491608 | 0.04 |
ENST00000545735.1 |
MANSC1 |
MANSC domain containing 1 |
| chr18_+_11751493 | 0.04 |
ENST00000269162.5 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr2_+_68961934 | 0.04 |
ENST00000409202.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr13_-_52703187 | 0.04 |
ENST00000355568.4 |
NEK5 |
NIMA-related kinase 5 |
| chr19_-_9649253 | 0.03 |
ENST00000593003.1 |
ZNF426 |
zinc finger protein 426 |
| chr16_+_810728 | 0.03 |
ENST00000563941.1 ENST00000545450.2 ENST00000566549.1 |
MSLN |
mesothelin |
| chr19_-_9649303 | 0.03 |
ENST00000253115.2 |
ZNF426 |
zinc finger protein 426 |
| chr1_-_16763911 | 0.03 |
ENST00000375577.1 ENST00000335496.1 |
SPATA21 |
spermatogenesis associated 21 |
| chr3_+_37493610 | 0.03 |
ENST00000264741.5 |
ITGA9 |
integrin, alpha 9 |
| chr1_-_155232221 | 0.03 |
ENST00000355379.3 |
SCAMP3 |
secretory carrier membrane protein 3 |
| chr11_-_57148619 | 0.02 |
ENST00000287143.2 |
PRG3 |
proteoglycan 3 |
| chr3_-_10452359 | 0.02 |
ENST00000452124.1 |
ATP2B2 |
ATPase, Ca++ transporting, plasma membrane 2 |
| chr6_-_132939317 | 0.02 |
ENST00000275191.2 |
TAAR2 |
trace amine associated receptor 2 |
| chr11_+_33902189 | 0.02 |
ENST00000330381.2 |
AC132216.1 |
HCG1785179; PRO1787; Uncharacterized protein |
| chr16_+_23765948 | 0.02 |
ENST00000300113.2 |
CHP2 |
calcineurin-like EF-hand protein 2 |
| chr22_-_43539346 | 0.02 |
ENST00000327555.5 ENST00000290429.6 |
MCAT |
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chr5_+_7396141 | 0.02 |
ENST00000338316.4 |
ADCY2 |
adenylate cyclase 2 (brain) |
| chr1_-_33647267 | 0.01 |
ENST00000291416.5 |
TRIM62 |
tripartite motif containing 62 |
| chr6_+_28493666 | 0.01 |
ENST00000412168.2 |
GPX5 |
glutathione peroxidase 5 (epididymal androgen-related protein) |
| chr17_+_79495397 | 0.01 |
ENST00000417245.2 ENST00000334850.7 |
FSCN2 |
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr11_-_64889529 | 0.01 |
ENST00000531743.1 ENST00000527548.1 ENST00000526555.1 ENST00000279259.3 |
FAU |
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr4_+_96761238 | 0.01 |
ENST00000295266.4 |
PDHA2 |
pyruvate dehydrogenase (lipoamide) alpha 2 |
| chr8_-_144679296 | 0.01 |
ENST00000317198.6 |
EEF1D |
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr1_-_148202536 | 0.01 |
ENST00000544708.1 |
PPIAL4D |
peptidylprolyl isomerase A (cyclophilin A)-like 4D |
| chr4_-_7105056 | 0.00 |
ENST00000504402.1 ENST00000499242.2 ENST00000501888.2 |
RP11-367J11.3 |
RP11-367J11.3 |
| chr16_+_78133293 | 0.00 |
ENST00000566780.1 |
WWOX |
WW domain containing oxidoreductase |
| chr2_-_85895295 | 0.00 |
ENST00000428225.1 ENST00000519937.2 |
SFTPB |
surfactant protein B |
| chr9_+_131447342 | 0.00 |
ENST00000409104.3 |
SET |
SET nuclear oncogene |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0044393 | microspike(GO:0044393) |
| 0.1 | 1.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 2.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 2.1 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 2.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1902948 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.4 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 0.4 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 1.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.3 | GO:0001757 | somite specification(GO:0001757) sclerotome development(GO:0061056) |
| 0.1 | 0.4 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.3 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 0.7 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0001188 | RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 1.0 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 2.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0090299 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.7 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 2.1 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |