Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
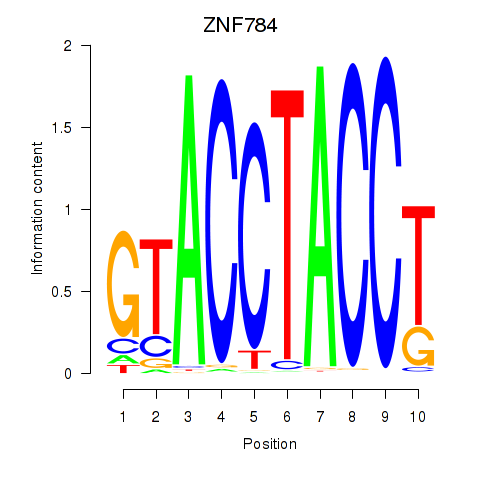
Results for ZNF784
Z-value: 1.00
Transcription factors associated with ZNF784
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF784
|
ENSG00000179922.5 | ZNF784 |
Activity profile of ZNF784 motif
Sorted Z-values of ZNF784 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF784
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_21529811 | 2.54 |
ENST00000588004.1 |
LAMA3 |
laminin, alpha 3 |
| chr12_-_52911718 | 2.47 |
ENST00000548409.1 |
KRT5 |
keratin 5 |
| chr4_-_90758227 | 1.86 |
ENST00000506691.1 ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr22_-_37640277 | 1.82 |
ENST00000401529.3 ENST00000249071.6 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr4_-_90758118 | 1.76 |
ENST00000420646.2 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr4_-_90756769 | 1.59 |
ENST00000345009.4 ENST00000505199.1 ENST00000502987.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr4_-_90757364 | 1.38 |
ENST00000508895.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr12_+_52626898 | 1.38 |
ENST00000331817.5 |
KRT7 |
keratin 7 |
| chr1_+_32042105 | 1.37 |
ENST00000457433.2 ENST00000441210.2 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
| chr1_+_32042131 | 1.35 |
ENST00000271064.7 ENST00000537531.1 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
| chr17_+_73717551 | 1.29 |
ENST00000450894.3 |
ITGB4 |
integrin, beta 4 |
| chr22_-_37640456 | 1.24 |
ENST00000405484.1 ENST00000441619.1 ENST00000406508.1 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr9_-_110251836 | 1.23 |
ENST00000374672.4 |
KLF4 |
Kruppel-like factor 4 (gut) |
| chr10_+_8096769 | 1.23 |
ENST00000346208.3 |
GATA3 |
GATA binding protein 3 |
| chr1_-_186649543 | 1.08 |
ENST00000367468.5 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr17_+_73717407 | 1.07 |
ENST00000579662.1 |
ITGB4 |
integrin, beta 4 |
| chr18_-_21852143 | 1.05 |
ENST00000399443.3 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr2_-_56150910 | 1.01 |
ENST00000424836.2 ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
| chr11_-_115375107 | 0.98 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chr15_-_59665062 | 0.98 |
ENST00000288235.4 |
MYO1E |
myosin IE |
| chr12_+_29376673 | 0.90 |
ENST00000547116.1 |
FAR2 |
fatty acyl CoA reductase 2 |
| chr10_+_8096631 | 0.89 |
ENST00000379328.3 |
GATA3 |
GATA binding protein 3 |
| chr12_+_29376592 | 0.87 |
ENST00000182377.4 |
FAR2 |
fatty acyl CoA reductase 2 |
| chr17_+_73717516 | 0.85 |
ENST00000200181.3 ENST00000339591.3 |
ITGB4 |
integrin, beta 4 |
| chr2_-_56150184 | 0.75 |
ENST00000394554.1 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
| chr20_+_44036620 | 0.75 |
ENST00000372710.3 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr6_+_139456226 | 0.74 |
ENST00000367658.2 |
HECA |
headcase homolog (Drosophila) |
| chr2_-_85636928 | 0.73 |
ENST00000449030.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr20_-_56286479 | 0.68 |
ENST00000265626.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr11_-_76381029 | 0.66 |
ENST00000407242.2 ENST00000421973.1 |
LRRC32 |
leucine rich repeat containing 32 |
| chr20_-_56285595 | 0.65 |
ENST00000395816.3 ENST00000347215.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr1_+_165600436 | 0.63 |
ENST00000367888.4 ENST00000367885.1 ENST00000367884.2 |
MGST3 |
microsomal glutathione S-transferase 3 |
| chr22_+_31003133 | 0.62 |
ENST00000405742.3 |
TCN2 |
transcobalamin II |
| chr5_-_1524015 | 0.61 |
ENST00000283415.3 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
| chr17_-_19281203 | 0.61 |
ENST00000487415.2 |
B9D1 |
B9 protein domain 1 |
| chr1_-_175161890 | 0.59 |
ENST00000545251.2 ENST00000423313.1 |
KIAA0040 |
KIAA0040 |
| chr18_+_61254534 | 0.59 |
ENST00000269489.5 |
SERPINB13 |
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr21_-_27542972 | 0.59 |
ENST00000346798.3 ENST00000439274.2 ENST00000354192.3 ENST00000348990.5 ENST00000357903.3 ENST00000358918.3 ENST00000359726.3 |
APP |
amyloid beta (A4) precursor protein |
| chr15_+_40531621 | 0.57 |
ENST00000560346.1 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr8_-_145018905 | 0.56 |
ENST00000398774.2 |
PLEC |
plectin |
| chr1_-_184943610 | 0.55 |
ENST00000367511.3 |
FAM129A |
family with sequence similarity 129, member A |
| chr12_+_49212514 | 0.54 |
ENST00000301050.2 ENST00000548279.1 ENST00000547230.1 |
CACNB3 |
calcium channel, voltage-dependent, beta 3 subunit |
| chr1_+_165600083 | 0.54 |
ENST00000367889.3 |
MGST3 |
microsomal glutathione S-transferase 3 |
| chr22_+_31002779 | 0.54 |
ENST00000215838.3 |
TCN2 |
transcobalamin II |
| chr6_-_46922659 | 0.53 |
ENST00000265417.7 |
GPR116 |
G protein-coupled receptor 116 |
| chr11_+_76494253 | 0.52 |
ENST00000333090.4 |
TSKU |
tsukushi, small leucine rich proteoglycan |
| chr20_+_44036900 | 0.49 |
ENST00000443296.1 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr17_+_42429493 | 0.47 |
ENST00000586242.1 |
GRN |
granulin |
| chrX_-_23761317 | 0.47 |
ENST00000492081.1 ENST00000379303.5 ENST00000336430.7 |
ACOT9 |
acyl-CoA thioesterase 9 |
| chr9_-_112260531 | 0.47 |
ENST00000374541.2 ENST00000262539.3 |
PTPN3 |
protein tyrosine phosphatase, non-receptor type 3 |
| chrY_+_16634483 | 0.46 |
ENST00000382872.1 |
NLGN4Y |
neuroligin 4, Y-linked |
| chr11_+_7506713 | 0.46 |
ENST00000329293.3 ENST00000534244.1 |
OLFML1 |
olfactomedin-like 1 |
| chr18_+_61254570 | 0.44 |
ENST00000344731.5 |
SERPINB13 |
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr16_-_31147020 | 0.43 |
ENST00000568261.1 ENST00000567797.1 ENST00000317508.6 |
PRSS8 |
protease, serine, 8 |
| chr9_+_116263639 | 0.42 |
ENST00000343817.5 |
RGS3 |
regulator of G-protein signaling 3 |
| chr9_+_116263778 | 0.41 |
ENST00000394646.3 |
RGS3 |
regulator of G-protein signaling 3 |
| chr3_+_197476621 | 0.41 |
ENST00000241502.4 |
FYTTD1 |
forty-two-three domain containing 1 |
| chr8_-_145016692 | 0.41 |
ENST00000357649.2 |
PLEC |
plectin |
| chr19_-_42498369 | 0.40 |
ENST00000302102.5 ENST00000545399.1 |
ATP1A3 |
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr7_-_107643674 | 0.39 |
ENST00000222399.6 |
LAMB1 |
laminin, beta 1 |
| chr19_-_42498231 | 0.38 |
ENST00000602133.1 |
ATP1A3 |
ATPase, Na+/K+ transporting, alpha 3 polypeptide |
| chr17_+_55055466 | 0.37 |
ENST00000262288.3 ENST00000572710.1 ENST00000575395.1 |
SCPEP1 |
serine carboxypeptidase 1 |
| chr3_-_122512619 | 0.36 |
ENST00000383659.1 ENST00000306103.2 |
HSPBAP1 |
HSPB (heat shock 27kDa) associated protein 1 |
| chr7_-_22234381 | 0.35 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr18_-_5419797 | 0.35 |
ENST00000542146.1 ENST00000427684.2 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
| chr11_-_62314268 | 0.35 |
ENST00000257247.7 ENST00000531324.1 ENST00000378024.4 |
AHNAK |
AHNAK nucleoprotein |
| chr2_-_11272234 | 0.34 |
ENST00000590207.1 ENST00000417697.2 ENST00000396164.1 ENST00000536743.1 ENST00000544306.1 |
AC062028.1 |
AC062028.1 |
| chr17_+_4736627 | 0.33 |
ENST00000355280.6 ENST00000347992.7 |
MINK1 |
misshapen-like kinase 1 |
| chrY_+_16636354 | 0.32 |
ENST00000339174.5 |
NLGN4Y |
neuroligin 4, Y-linked |
| chr3_-_42306248 | 0.32 |
ENST00000334681.5 |
CCK |
cholecystokinin |
| chr17_+_77020325 | 0.31 |
ENST00000311661.4 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
| chr17_+_77020224 | 0.31 |
ENST00000339142.2 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
| chr12_-_14996355 | 0.31 |
ENST00000228936.4 |
ART4 |
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr18_-_45457192 | 0.29 |
ENST00000586514.1 ENST00000591214.1 ENST00000589877.1 |
SMAD2 |
SMAD family member 2 |
| chr1_+_171283331 | 0.29 |
ENST00000367749.3 |
FMO4 |
flavin containing monooxygenase 4 |
| chr3_+_9834227 | 0.29 |
ENST00000287613.7 ENST00000397261.3 |
ARPC4 |
actin related protein 2/3 complex, subunit 4, 20kDa |
| chr2_-_25873079 | 0.28 |
ENST00000496972.2 |
DTNB |
dystrobrevin, beta |
| chr17_+_19281034 | 0.28 |
ENST00000308406.5 ENST00000299612.7 |
MAPK7 |
mitogen-activated protein kinase 7 |
| chr7_+_114562172 | 0.27 |
ENST00000393486.1 ENST00000257724.3 |
MDFIC |
MyoD family inhibitor domain containing |
| chr6_+_136172820 | 0.27 |
ENST00000308191.6 |
PDE7B |
phosphodiesterase 7B |
| chr2_-_70780572 | 0.27 |
ENST00000450929.1 |
TGFA |
transforming growth factor, alpha |
| chr3_+_16926441 | 0.27 |
ENST00000418129.2 ENST00000396755.2 |
PLCL2 |
phospholipase C-like 2 |
| chr17_-_56494713 | 0.26 |
ENST00000407977.2 |
RNF43 |
ring finger protein 43 |
| chr1_-_153029980 | 0.26 |
ENST00000392653.2 |
SPRR2A |
small proline-rich protein 2A |
| chr11_-_36619771 | 0.25 |
ENST00000311485.3 ENST00000527033.1 ENST00000532616.1 |
RAG2 |
recombination activating gene 2 |
| chr1_-_207095324 | 0.24 |
ENST00000530505.1 ENST00000367091.3 ENST00000442471.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr16_-_67190152 | 0.24 |
ENST00000486556.1 |
TRADD |
TNFRSF1A-associated via death domain |
| chr5_+_137225125 | 0.24 |
ENST00000350250.4 ENST00000508638.1 ENST00000502810.1 ENST00000508883.1 |
PKD2L2 |
polycystic kidney disease 2-like 2 |
| chr12_+_52445191 | 0.24 |
ENST00000243050.1 ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1 |
nuclear receptor subfamily 4, group A, member 1 |
| chr1_+_228327923 | 0.23 |
ENST00000391865.3 |
GUK1 |
guanylate kinase 1 |
| chr18_-_45456930 | 0.22 |
ENST00000262160.6 ENST00000587269.1 |
SMAD2 |
SMAD family member 2 |
| chr17_-_1394940 | 0.22 |
ENST00000570984.2 ENST00000361007.2 |
MYO1C |
myosin IC |
| chr15_+_25200108 | 0.21 |
ENST00000577949.1 ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF SNRPN |
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr11_+_125439298 | 0.21 |
ENST00000278903.6 ENST00000343678.4 ENST00000524723.1 ENST00000527842.2 |
EI24 |
etoposide induced 2.4 |
| chr22_+_23522552 | 0.20 |
ENST00000359540.3 ENST00000398512.5 |
BCR |
breakpoint cluster region |
| chr1_+_90287480 | 0.20 |
ENST00000394593.3 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
| chr11_+_129939811 | 0.20 |
ENST00000345598.5 ENST00000338167.5 |
APLP2 |
amyloid beta (A4) precursor-like protein 2 |
| chr11_+_129939779 | 0.20 |
ENST00000533195.1 ENST00000533713.1 ENST00000528499.1 ENST00000539648.1 ENST00000263574.5 |
APLP2 |
amyloid beta (A4) precursor-like protein 2 |
| chr15_+_40532058 | 0.20 |
ENST00000260404.4 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr1_-_207095212 | 0.19 |
ENST00000420007.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr17_-_56494882 | 0.19 |
ENST00000584437.1 |
RNF43 |
ring finger protein 43 |
| chr7_+_26332645 | 0.19 |
ENST00000396376.1 |
SNX10 |
sorting nexin 10 |
| chr17_+_1944790 | 0.18 |
ENST00000575162.1 |
DPH1 |
diphthamide biosynthesis 1 |
| chr17_-_56494908 | 0.18 |
ENST00000577716.1 |
RNF43 |
ring finger protein 43 |
| chr19_-_14629224 | 0.18 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr1_+_228327943 | 0.18 |
ENST00000366726.1 ENST00000312726.4 ENST00000366728.2 ENST00000453943.1 ENST00000366723.1 ENST00000366722.1 ENST00000435153.1 ENST00000366721.1 |
GUK1 |
guanylate kinase 1 |
| chr19_+_50879705 | 0.18 |
ENST00000598168.1 ENST00000411902.2 ENST00000253727.5 ENST00000597790.1 ENST00000597130.1 ENST00000599105.1 |
NR1H2 |
nuclear receptor subfamily 1, group H, member 2 |
| chr3_+_9834179 | 0.18 |
ENST00000498623.2 |
ARPC4 |
actin related protein 2/3 complex, subunit 4, 20kDa |
| chr1_+_50574585 | 0.17 |
ENST00000371824.1 ENST00000371823.4 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr3_+_136581096 | 0.17 |
ENST00000476286.1 ENST00000488930.1 |
NCK1 |
NCK adaptor protein 1 |
| chr16_+_2564254 | 0.17 |
ENST00000565223.1 |
ATP6V0C |
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
| chr1_+_47901689 | 0.17 |
ENST00000334793.5 |
FOXD2 |
forkhead box D2 |
| chr15_+_71839566 | 0.17 |
ENST00000357769.4 |
THSD4 |
thrombospondin, type I, domain containing 4 |
| chrX_+_106045891 | 0.16 |
ENST00000357242.5 ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B |
TBC1 domain family, member 8B (with GRAM domain) |
| chr1_+_203651937 | 0.16 |
ENST00000341360.2 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
| chr12_+_57828521 | 0.16 |
ENST00000309668.2 |
INHBC |
inhibin, beta C |
| chr2_+_173940668 | 0.15 |
ENST00000375213.3 |
MLTK |
Mitogen-activated protein kinase kinase kinase MLT |
| chr6_-_56258892 | 0.15 |
ENST00000370819.1 |
COL21A1 |
collagen, type XXI, alpha 1 |
| chr3_+_45067659 | 0.15 |
ENST00000296130.4 |
CLEC3B |
C-type lectin domain family 3, member B |
| chr3_-_121379739 | 0.14 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr14_+_105452094 | 0.14 |
ENST00000551606.1 ENST00000547315.1 |
C14orf79 |
chromosome 14 open reading frame 79 |
| chr1_-_115238207 | 0.14 |
ENST00000520113.2 ENST00000369538.3 ENST00000353928.6 |
AMPD1 |
adenosine monophosphate deaminase 1 |
| chr19_+_13858593 | 0.14 |
ENST00000221554.8 |
CCDC130 |
coiled-coil domain containing 130 |
| chr17_+_1933404 | 0.14 |
ENST00000263083.6 ENST00000571418.1 |
DPH1 |
diphthamide biosynthesis 1 |
| chr3_-_48130707 | 0.14 |
ENST00000360240.6 ENST00000383737.4 |
MAP4 |
microtubule-associated protein 4 |
| chr5_-_74062930 | 0.13 |
ENST00000509430.1 ENST00000345239.2 ENST00000427854.2 ENST00000506778.1 |
GFM2 |
G elongation factor, mitochondrial 2 |
| chr16_+_19179549 | 0.13 |
ENST00000355377.2 ENST00000568115.1 |
SYT17 |
synaptotagmin XVII |
| chr2_-_70780770 | 0.13 |
ENST00000444975.1 ENST00000445399.1 ENST00000418333.2 |
TGFA |
transforming growth factor, alpha |
| chr11_-_6440624 | 0.13 |
ENST00000311051.3 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr11_-_6440283 | 0.13 |
ENST00000299402.6 ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr7_+_119913688 | 0.13 |
ENST00000331113.4 |
KCND2 |
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr3_-_195310802 | 0.13 |
ENST00000421243.1 ENST00000453131.1 |
APOD |
apolipoprotein D |
| chr12_-_58131931 | 0.13 |
ENST00000547588.1 |
AGAP2 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr4_+_158141899 | 0.12 |
ENST00000264426.9 ENST00000506284.1 |
GRIA2 |
glutamate receptor, ionotropic, AMPA 2 |
| chrX_-_106243451 | 0.12 |
ENST00000355610.4 ENST00000535534.1 |
MORC4 |
MORC family CW-type zinc finger 4 |
| chr5_+_137225158 | 0.12 |
ENST00000290431.5 |
PKD2L2 |
polycystic kidney disease 2-like 2 |
| chr9_-_127533519 | 0.11 |
ENST00000487099.2 ENST00000344523.4 ENST00000373584.3 |
NR6A1 |
nuclear receptor subfamily 6, group A, member 1 |
| chr4_-_153274078 | 0.11 |
ENST00000263981.5 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr6_-_45983581 | 0.11 |
ENST00000339561.6 |
CLIC5 |
chloride intracellular channel 5 |
| chr20_+_20348740 | 0.11 |
ENST00000310227.1 |
INSM1 |
insulinoma-associated 1 |
| chr6_-_47009996 | 0.10 |
ENST00000371243.2 |
GPR110 |
G protein-coupled receptor 110 |
| chr3_-_48130314 | 0.10 |
ENST00000439356.1 ENST00000395734.3 ENST00000426837.2 |
MAP4 |
microtubule-associated protein 4 |
| chr5_+_140729649 | 0.10 |
ENST00000523390.1 |
PCDHGB1 |
protocadherin gamma subfamily B, 1 |
| chr8_-_16859690 | 0.10 |
ENST00000180166.5 |
FGF20 |
fibroblast growth factor 20 |
| chr5_-_159827033 | 0.10 |
ENST00000523213.1 |
C5orf54 |
chromosome 5 open reading frame 54 |
| chr11_+_3829691 | 0.10 |
ENST00000278243.4 ENST00000463452.2 ENST00000479072.1 ENST00000496834.2 ENST00000469307.2 |
PGAP2 |
post-GPI attachment to proteins 2 |
| chr12_+_54402790 | 0.09 |
ENST00000040584.4 |
HOXC8 |
homeobox C8 |
| chr5_-_159827073 | 0.09 |
ENST00000408953.3 |
C5orf54 |
chromosome 5 open reading frame 54 |
| chr22_+_42196666 | 0.09 |
ENST00000402061.3 ENST00000255784.5 |
CCDC134 |
coiled-coil domain containing 134 |
| chr11_+_31531291 | 0.09 |
ENST00000350638.5 ENST00000379163.5 ENST00000395934.2 |
ELP4 |
elongator acetyltransferase complex subunit 4 |
| chr2_+_45878790 | 0.08 |
ENST00000306156.3 |
PRKCE |
protein kinase C, epsilon |
| chr2_+_226265364 | 0.08 |
ENST00000272907.6 |
NYAP2 |
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr11_-_19262486 | 0.08 |
ENST00000250024.4 |
E2F8 |
E2F transcription factor 8 |
| chr12_-_66524482 | 0.08 |
ENST00000446587.2 ENST00000266604.2 |
LLPH |
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chrX_+_101975643 | 0.08 |
ENST00000361229.4 |
BHLHB9 |
basic helix-loop-helix domain containing, class B, 9 |
| chrX_+_101975619 | 0.07 |
ENST00000457056.1 |
BHLHB9 |
basic helix-loop-helix domain containing, class B, 9 |
| chr4_+_158142750 | 0.07 |
ENST00000505888.1 ENST00000449365.1 |
GRIA2 |
glutamate receptor, ionotropic, AMPA 2 |
| chr8_+_85095497 | 0.07 |
ENST00000522455.1 ENST00000521695.1 |
RALYL |
RALY RNA binding protein-like |
| chr7_-_558876 | 0.07 |
ENST00000354513.5 ENST00000402802.3 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
| chr11_+_64008525 | 0.07 |
ENST00000449942.2 |
FKBP2 |
FK506 binding protein 2, 13kDa |
| chr11_+_64008443 | 0.07 |
ENST00000309366.4 |
FKBP2 |
FK506 binding protein 2, 13kDa |
| chr17_-_74707037 | 0.07 |
ENST00000355797.3 ENST00000375036.2 ENST00000449428.2 |
MXRA7 |
matrix-remodelling associated 7 |
| chr12_+_7060432 | 0.07 |
ENST00000318974.9 ENST00000456013.1 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr1_+_54411715 | 0.06 |
ENST00000371370.3 ENST00000371368.1 |
LRRC42 |
leucine rich repeat containing 42 |
| chr1_+_115572415 | 0.06 |
ENST00000256592.1 |
TSHB |
thyroid stimulating hormone, beta |
| chr2_-_24583583 | 0.06 |
ENST00000355123.4 |
ITSN2 |
intersectin 2 |
| chr12_-_4754339 | 0.06 |
ENST00000228850.1 |
AKAP3 |
A kinase (PRKA) anchor protein 3 |
| chr11_-_8290263 | 0.06 |
ENST00000428101.2 |
LMO1 |
LIM domain only 1 (rhombotin 1) |
| chr12_+_14518598 | 0.06 |
ENST00000261168.4 ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP |
activating transcription factor 7 interacting protein |
| chr9_+_132815985 | 0.05 |
ENST00000372410.3 |
GPR107 |
G protein-coupled receptor 107 |
| chr9_+_130565487 | 0.05 |
ENST00000373225.3 ENST00000431857.1 |
FPGS |
folylpolyglutamate synthase |
| chr12_+_53895364 | 0.05 |
ENST00000552817.1 ENST00000394357.2 |
TARBP2 |
TAR (HIV-1) RNA binding protein 2 |
| chr8_+_80523321 | 0.05 |
ENST00000518111.1 |
STMN2 |
stathmin-like 2 |
| chr9_+_136325089 | 0.05 |
ENST00000291722.7 ENST00000316948.4 ENST00000540581.1 |
CACFD1 |
calcium channel flower domain containing 1 |
| chr3_+_184038073 | 0.05 |
ENST00000428387.1 ENST00000434061.2 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr11_+_86013253 | 0.04 |
ENST00000533986.1 ENST00000278483.3 |
C11orf73 |
chromosome 11 open reading frame 73 |
| chr7_+_22766766 | 0.04 |
ENST00000426291.1 ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6 |
interleukin 6 (interferon, beta 2) |
| chr12_+_57998400 | 0.04 |
ENST00000548804.1 ENST00000550596.1 ENST00000551835.1 ENST00000549583.1 |
DTX3 |
deltex homolog 3 (Drosophila) |
| chr19_+_3880581 | 0.04 |
ENST00000450849.2 ENST00000301260.6 ENST00000398448.3 |
ATCAY |
ataxia, cerebellar, Cayman type |
| chr11_-_10590238 | 0.04 |
ENST00000256178.3 |
LYVE1 |
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr8_-_57358432 | 0.03 |
ENST00000517415.1 ENST00000314922.3 |
PENK |
proenkephalin |
| chr11_-_73720122 | 0.03 |
ENST00000426995.2 |
UCP3 |
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr12_-_12837423 | 0.03 |
ENST00000540510.1 |
GPR19 |
G protein-coupled receptor 19 |
| chr11_-_10590118 | 0.03 |
ENST00000529598.1 |
LYVE1 |
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr17_-_50237343 | 0.03 |
ENST00000575181.1 ENST00000570565.1 |
CA10 |
carbonic anhydrase X |
| chr11_-_67271723 | 0.03 |
ENST00000533391.1 ENST00000534749.1 ENST00000532703.1 |
PITPNM1 |
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr5_+_161274940 | 0.03 |
ENST00000393943.4 |
GABRA1 |
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr11_+_65154070 | 0.02 |
ENST00000317568.5 ENST00000531296.1 ENST00000533782.1 ENST00000355991.5 ENST00000416776.2 ENST00000526201.1 |
FRMD8 |
FERM domain containing 8 |
| chr19_+_39936186 | 0.02 |
ENST00000432763.2 ENST00000402194.2 ENST00000601515.1 |
SUPT5H |
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr1_-_92951607 | 0.02 |
ENST00000427103.1 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr22_-_44708731 | 0.02 |
ENST00000381176.4 |
KIAA1644 |
KIAA1644 |
| chr9_+_88556036 | 0.02 |
ENST00000361671.5 ENST00000416045.1 |
NAA35 |
N(alpha)-acetyltransferase 35, NatC auxiliary subunit |
| chr1_+_20465805 | 0.02 |
ENST00000375102.3 |
PLA2G2F |
phospholipase A2, group IIF |
| chr4_+_71248795 | 0.02 |
ENST00000304915.3 |
SMR3B |
submaxillary gland androgen regulated protein 3B |
| chr19_+_39936317 | 0.02 |
ENST00000598725.1 |
SUPT5H |
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr4_+_48833119 | 0.02 |
ENST00000444354.2 ENST00000509963.1 ENST00000509246.1 |
OCIAD1 |
OCIA domain containing 1 |
| chr3_+_32993065 | 0.02 |
ENST00000330953.5 |
CCR4 |
chemokine (C-C motif) receptor 4 |
| chr1_+_205012293 | 0.02 |
ENST00000331830.4 |
CNTN2 |
contactin 2 (axonal) |
| chr4_-_187476721 | 0.02 |
ENST00000307161.5 |
MTNR1A |
melatonin receptor 1A |
| chr4_-_130692631 | 0.01 |
ENST00000500092.2 ENST00000509105.1 |
RP11-519M16.1 |
RP11-519M16.1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.6 | GO:2000468 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) regulation of peroxidase activity(GO:2000468) positive regulation of peroxidase activity(GO:2000470) |
| 1.1 | 1.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.7 | 2.1 | GO:1901536 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.6 | 1.8 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.4 | 1.2 | GO:0071409 | negative regulation of muscle hyperplasia(GO:0014740) cellular response to cycloheximide(GO:0071409) |
| 0.3 | 9.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 3.0 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.2 | 1.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.2 | 0.7 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.2 | 1.8 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.2 | 0.5 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.2 | 0.5 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.2 | 1.0 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.2 | 0.6 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 0.6 | GO:2000857 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.4 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) dGDP metabolic process(GO:0046066) |
| 0.1 | 0.6 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 1.2 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 1.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.8 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 1.0 | GO:0097283 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.1 | 0.3 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 0.3 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.2 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.2 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 0.2 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.2 | GO:1900082 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.1 | 0.5 | GO:0098902 | regulation of membrane depolarization during action potential(GO:0098902) |
| 0.0 | 0.3 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.3 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.5 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.0 | 0.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.2 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.7 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 1.2 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.2 | GO:0061469 | response to corticotropin-releasing hormone(GO:0043435) regulation of type B pancreatic cell proliferation(GO:0061469) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 1.1 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.3 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0006525 | arginine metabolic process(GO:0006525) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.4 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 1.3 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 6.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 2.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.8 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.8 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 4.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 7.0 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 4.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.4 | GO:0043259 | laminin-2 complex(GO:0005607) laminin-10 complex(GO:0043259) |
| 0.1 | 0.9 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 0.8 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 3.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.5 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.1 | 1.8 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.2 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) Flemming body(GO:0090543) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 3.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0098843 | postsynaptic endocytic zone(GO:0098843) |
| 0.0 | 1.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.3 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 1.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 1.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 1.2 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 1.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.6 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.6 | 1.8 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.4 | 2.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.4 | 1.8 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.3 | 1.4 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.3 | 1.1 | GO:0050473 | prostaglandin-endoperoxide synthase activity(GO:0004666) arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.2 | 3.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.2 | 0.6 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 2.7 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.6 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.6 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.8 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 0.2 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 1.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.5 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.1 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 1.7 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 1.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 3.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 7.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 1.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |