Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for ZSCAN4
Z-value: 0.88
Transcription factors associated with ZSCAN4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
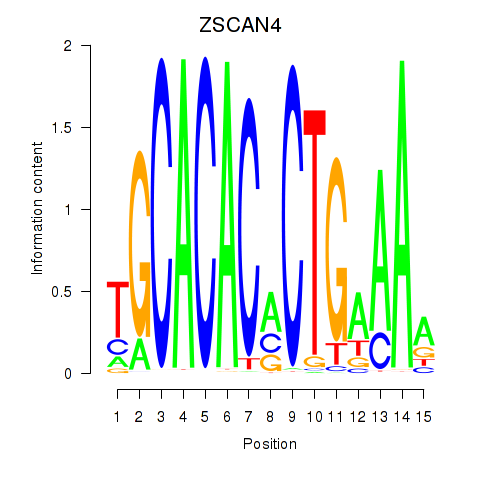
ZSCAN4
|
ENSG00000180532.6 | ZSCAN4 |
Activity profile of ZSCAN4 motif
Sorted Z-values of ZSCAN4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ZSCAN4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_49160148 | 3.51 |
ENST00000407599.3 |
GAGE10 |
G antigen 10 |
| chr19_-_17516449 | 2.26 |
ENST00000252593.6 |
BST2 |
bone marrow stromal cell antigen 2 |
| chr11_-_75062730 | 1.98 |
ENST00000420843.2 ENST00000360025.3 |
ARRB1 |
arrestin, beta 1 |
| chr12_-_54694807 | 1.65 |
ENST00000435572.2 |
NFE2 |
nuclear factor, erythroid 2 |
| chr16_+_23847267 | 1.46 |
ENST00000321728.7 |
PRKCB |
protein kinase C, beta |
| chr2_+_189156389 | 1.22 |
ENST00000409843.1 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
| chr14_-_21493884 | 1.20 |
ENST00000556974.1 ENST00000554419.1 ENST00000298687.5 ENST00000397858.1 ENST00000360463.3 ENST00000350792.3 ENST00000397847.2 |
NDRG2 |
NDRG family member 2 |
| chr14_-_21493649 | 1.17 |
ENST00000553442.1 ENST00000555869.1 ENST00000556457.1 ENST00000397844.2 ENST00000554415.1 |
NDRG2 |
NDRG family member 2 |
| chr22_-_24384260 | 1.05 |
ENST00000248935.5 |
GSTT1 |
glutathione S-transferase theta 1 |
| chr3_-_52569023 | 1.01 |
ENST00000307076.4 |
NT5DC2 |
5'-nucleotidase domain containing 2 |
| chr21_-_33651324 | 0.86 |
ENST00000290130.3 |
MIS18A |
MIS18 kinetochore protein A |
| chr9_+_125137565 | 0.86 |
ENST00000373698.5 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr1_-_21978312 | 0.84 |
ENST00000359708.4 ENST00000290101.4 |
RAP1GAP |
RAP1 GTPase activating protein |
| chr2_-_101034070 | 0.83 |
ENST00000264249.3 |
CHST10 |
carbohydrate sulfotransferase 10 |
| chr16_+_29690358 | 0.77 |
ENST00000395384.4 ENST00000562473.1 |
QPRT |
quinolinate phosphoribosyltransferase |
| chr17_-_7080227 | 0.73 |
ENST00000574330.1 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr21_-_46221684 | 0.68 |
ENST00000330942.5 |
UBE2G2 |
ubiquitin-conjugating enzyme E2G 2 |
| chr15_-_37392703 | 0.64 |
ENST00000382766.2 ENST00000444725.1 |
MEIS2 |
Meis homeobox 2 |
| chr14_-_55658252 | 0.64 |
ENST00000395425.2 |
DLGAP5 |
discs, large (Drosophila) homolog-associated protein 5 |
| chr6_-_41909466 | 0.59 |
ENST00000414200.2 |
CCND3 |
cyclin D3 |
| chr1_+_52195542 | 0.56 |
ENST00000462759.1 ENST00000486942.1 |
OSBPL9 |
oxysterol binding protein-like 9 |
| chr1_+_233765353 | 0.55 |
ENST00000366620.1 |
KCNK1 |
potassium channel, subfamily K, member 1 |
| chr6_-_153304697 | 0.52 |
ENST00000367241.3 |
FBXO5 |
F-box protein 5 |
| chr9_+_139557360 | 0.51 |
ENST00000308874.7 ENST00000406555.3 ENST00000492862.2 |
EGFL7 |
EGF-like-domain, multiple 7 |
| chr19_-_3801789 | 0.51 |
ENST00000590849.1 ENST00000395045.2 |
MATK |
megakaryocyte-associated tyrosine kinase |
| chr11_+_66624527 | 0.50 |
ENST00000393952.3 |
LRFN4 |
leucine rich repeat and fibronectin type III domain containing 4 |
| chr11_+_111807863 | 0.49 |
ENST00000440460.2 |
DIXDC1 |
DIX domain containing 1 |
| chr6_-_41909561 | 0.49 |
ENST00000372991.4 |
CCND3 |
cyclin D3 |
| chr5_+_176784837 | 0.48 |
ENST00000408923.3 |
RGS14 |
regulator of G-protein signaling 14 |
| chr22_+_39853258 | 0.45 |
ENST00000341184.6 |
MGAT3 |
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr19_-_3025614 | 0.40 |
ENST00000447365.2 |
TLE2 |
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr2_+_183580954 | 0.38 |
ENST00000264065.7 |
DNAJC10 |
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr5_+_61602055 | 0.38 |
ENST00000381103.2 |
KIF2A |
kinesin heavy chain member 2A |
| chr10_-_62332357 | 0.36 |
ENST00000503366.1 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr10_-_101989315 | 0.36 |
ENST00000370397.7 |
CHUK |
conserved helix-loop-helix ubiquitous kinase |
| chr10_-_30348439 | 0.36 |
ENST00000375377.1 |
KIAA1462 |
KIAA1462 |
| chr10_-_103880209 | 0.35 |
ENST00000425280.1 |
LDB1 |
LIM domain binding 1 |
| chr3_-_53290016 | 0.34 |
ENST00000423525.2 ENST00000423516.1 ENST00000296289.6 ENST00000462138.1 |
TKT |
transketolase |
| chrX_-_133792480 | 0.34 |
ENST00000359237.4 |
PLAC1 |
placenta-specific 1 |
| chr3_+_98482175 | 0.33 |
ENST00000485391.1 ENST00000492254.1 |
ST3GAL6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr6_-_76203345 | 0.33 |
ENST00000393004.2 |
FILIP1 |
filamin A interacting protein 1 |
| chr15_+_63414017 | 0.33 |
ENST00000413507.2 |
LACTB |
lactamase, beta |
| chr8_+_107670064 | 0.31 |
ENST00000312046.6 |
OXR1 |
oxidation resistance 1 |
| chr19_+_17516531 | 0.31 |
ENST00000528911.1 ENST00000528604.1 ENST00000595892.1 ENST00000500836.2 ENST00000598546.1 ENST00000600369.1 ENST00000598356.1 ENST00000594426.1 |
MVB12A CTD-2521M24.9 |
multivesicular body subunit 12A CTD-2521M24.9 |
| chr7_+_107224364 | 0.30 |
ENST00000491150.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr12_+_122459757 | 0.29 |
ENST00000261822.4 |
BCL7A |
B-cell CLL/lymphoma 7A |
| chr19_+_17516494 | 0.28 |
ENST00000534306.1 |
CTD-2521M24.9 |
CTD-2521M24.9 |
| chr11_+_125462690 | 0.27 |
ENST00000392708.4 ENST00000529196.1 ENST00000531491.1 |
STT3A |
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr6_-_31620149 | 0.27 |
ENST00000435080.1 ENST00000375976.4 ENST00000441054.1 |
BAG6 |
BCL2-associated athanogene 6 |
| chr4_-_168155169 | 0.27 |
ENST00000534949.1 ENST00000535728.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr12_+_122242597 | 0.26 |
ENST00000267197.5 |
SETD1B |
SET domain containing 1B |
| chr9_+_103790991 | 0.25 |
ENST00000374874.3 |
LPPR1 |
Lipid phosphate phosphatase-related protein type 1 |
| chr20_+_57875658 | 0.25 |
ENST00000371025.3 |
EDN3 |
endothelin 3 |
| chr1_+_159409512 | 0.24 |
ENST00000423932.3 |
OR10J1 |
olfactory receptor, family 10, subfamily J, member 1 |
| chr7_-_10979750 | 0.24 |
ENST00000339600.5 |
NDUFA4 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa |
| chr1_+_226736446 | 0.24 |
ENST00000366788.3 ENST00000366789.4 |
C1orf95 |
chromosome 1 open reading frame 95 |
| chr1_-_161600990 | 0.23 |
ENST00000531221.1 |
FCGR3B |
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
| chr13_-_52733980 | 0.23 |
ENST00000339406.3 |
NEK3 |
NIMA-related kinase 3 |
| chr19_+_44716678 | 0.22 |
ENST00000586228.1 ENST00000588219.1 ENST00000313040.7 ENST00000589707.1 ENST00000588394.1 ENST00000589005.1 |
ZNF227 |
zinc finger protein 227 |
| chr17_+_9066252 | 0.22 |
ENST00000436734.1 |
NTN1 |
netrin 1 |
| chr7_-_99063769 | 0.22 |
ENST00000394186.3 ENST00000359832.4 ENST00000449683.1 ENST00000488775.1 ENST00000523680.1 ENST00000292475.3 ENST00000430982.1 ENST00000555673.1 ENST00000413834.1 |
ATP5J2 PTCD1 ATP5J2-PTCD1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F2 pentatricopeptide repeat domain 1 ATP5J2-PTCD1 readthrough |
| chr1_-_171621815 | 0.22 |
ENST00000037502.6 |
MYOC |
myocilin, trabecular meshwork inducible glucocorticoid response |
| chr19_-_38714847 | 0.21 |
ENST00000420980.2 ENST00000355526.4 |
DPF1 |
D4, zinc and double PHD fingers family 1 |
| chr19_+_14491948 | 0.21 |
ENST00000358600.3 |
CD97 |
CD97 molecule |
| chr2_-_70520539 | 0.21 |
ENST00000482975.2 ENST00000438261.1 |
SNRPG |
small nuclear ribonucleoprotein polypeptide G |
| chr15_-_41836441 | 0.20 |
ENST00000567866.1 ENST00000561603.1 ENST00000304330.4 ENST00000566863.1 |
RPAP1 |
RNA polymerase II associated protein 1 |
| chr4_-_168155417 | 0.18 |
ENST00000511269.1 ENST00000506697.1 ENST00000512042.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr4_-_152682129 | 0.18 |
ENST00000512306.1 ENST00000508611.1 ENST00000515812.1 ENST00000263985.6 |
PET112 |
PET112 homolog (yeast) |
| chr19_+_17516624 | 0.17 |
ENST00000596322.1 ENST00000600008.1 ENST00000601885.1 |
CTD-2521M24.9 |
CTD-2521M24.9 |
| chr1_-_161600822 | 0.17 |
ENST00000534776.1 ENST00000540048.1 |
FCGR3B FCGR3A |
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr1_-_161600942 | 0.17 |
ENST00000421702.2 |
FCGR3B |
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
| chr6_-_33548006 | 0.17 |
ENST00000374467.3 |
BAK1 |
BCL2-antagonist/killer 1 |
| chr10_+_133753533 | 0.17 |
ENST00000422256.2 |
PPP2R2D |
protein phosphatase 2, regulatory subunit B, delta |
| chr14_+_53173910 | 0.16 |
ENST00000606149.1 ENST00000555339.1 ENST00000556813.1 |
PSMC6 |
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr22_+_17082732 | 0.15 |
ENST00000558085.2 ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1 |
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr17_-_73761222 | 0.15 |
ENST00000437911.1 ENST00000225614.2 |
GALK1 |
galactokinase 1 |
| chr6_-_42690312 | 0.15 |
ENST00000230381.5 |
PRPH2 |
peripherin 2 (retinal degeneration, slow) |
| chr1_+_104615595 | 0.15 |
ENST00000418362.1 |
RP11-364B6.1 |
RP11-364B6.1 |
| chr16_+_4674814 | 0.14 |
ENST00000415496.1 ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr2_-_239198743 | 0.14 |
ENST00000440245.1 ENST00000431832.1 |
PER2 |
period circadian clock 2 |
| chr14_+_53173890 | 0.14 |
ENST00000445930.2 |
PSMC6 |
proteasome (prosome, macropain) 26S subunit, ATPase, 6 |
| chr16_+_15737124 | 0.14 |
ENST00000396355.1 ENST00000396353.2 |
NDE1 |
nudE neurodevelopment protein 1 |
| chr20_-_60573188 | 0.13 |
ENST00000474089.1 |
TAF4 |
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa |
| chr19_+_30863271 | 0.13 |
ENST00000355537.3 |
ZNF536 |
zinc finger protein 536 |
| chr19_-_42636617 | 0.13 |
ENST00000529067.1 ENST00000529952.1 ENST00000533720.1 ENST00000389341.5 ENST00000342301.4 |
POU2F2 |
POU class 2 homeobox 2 |
| chr2_-_167232484 | 0.13 |
ENST00000375387.4 ENST00000303354.6 ENST00000409672.1 |
SCN9A |
sodium channel, voltage-gated, type IX, alpha subunit |
| chr10_+_18549645 | 0.13 |
ENST00000396576.2 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr4_-_168155577 | 0.13 |
ENST00000541354.1 ENST00000509854.1 ENST00000512681.1 ENST00000421836.2 ENST00000510741.1 ENST00000510403.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr2_-_214016314 | 0.13 |
ENST00000434687.1 ENST00000374319.4 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
| chr1_+_159141397 | 0.13 |
ENST00000368124.4 ENST00000368125.4 ENST00000416746.1 |
CADM3 |
cell adhesion molecule 3 |
| chr20_-_16554078 | 0.12 |
ENST00000354981.2 ENST00000355755.3 ENST00000378003.2 ENST00000408042.1 |
KIF16B |
kinesin family member 16B |
| chr17_+_9728828 | 0.12 |
ENST00000262441.5 |
GLP2R |
glucagon-like peptide 2 receptor |
| chr9_+_107526443 | 0.12 |
ENST00000374762.3 |
NIPSNAP3B |
nipsnap homolog 3B (C. elegans) |
| chr20_-_21494654 | 0.11 |
ENST00000377142.4 |
NKX2-2 |
NK2 homeobox 2 |
| chr4_+_15779901 | 0.11 |
ENST00000226279.3 |
CD38 |
CD38 molecule |
| chr13_-_99404875 | 0.11 |
ENST00000376503.5 |
SLC15A1 |
solute carrier family 15 (oligopeptide transporter), member 1 |
| chr17_-_40337470 | 0.10 |
ENST00000293330.1 |
HCRT |
hypocretin (orexin) neuropeptide precursor |
| chr19_-_10530784 | 0.10 |
ENST00000593124.1 |
CDC37 |
cell division cycle 37 |
| chr4_-_168155300 | 0.10 |
ENST00000541637.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr13_+_60971080 | 0.10 |
ENST00000377894.2 |
TDRD3 |
tudor domain containing 3 |
| chr17_-_19648683 | 0.09 |
ENST00000573368.1 ENST00000457500.2 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
| chr13_+_60971427 | 0.09 |
ENST00000535286.1 ENST00000377881.2 |
TDRD3 |
tudor domain containing 3 |
| chr17_-_34207295 | 0.08 |
ENST00000463941.1 ENST00000293272.3 |
CCL5 |
chemokine (C-C motif) ligand 5 |
| chr2_+_220436917 | 0.08 |
ENST00000243786.2 |
INHA |
inhibin, alpha |
| chr18_+_6729698 | 0.08 |
ENST00000383472.4 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr1_-_193075180 | 0.08 |
ENST00000367440.3 |
GLRX2 |
glutaredoxin 2 |
| chr17_-_56406117 | 0.08 |
ENST00000268893.6 ENST00000355701.3 |
BZRAP1 |
benzodiazepine receptor (peripheral) associated protein 1 |
| chr11_-_33774944 | 0.07 |
ENST00000532057.1 ENST00000531080.1 |
FBXO3 |
F-box protein 3 |
| chr15_-_74374891 | 0.07 |
ENST00000290438.3 |
GOLGA6A |
golgin A6 family, member A |
| chr6_+_90272027 | 0.07 |
ENST00000522441.1 |
ANKRD6 |
ankyrin repeat domain 6 |
| chr18_+_32173276 | 0.07 |
ENST00000591816.1 ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA |
dystrobrevin, alpha |
| chr19_-_42636543 | 0.07 |
ENST00000528894.4 ENST00000560804.2 ENST00000560558.1 ENST00000560398.1 ENST00000526816.2 |
POU2F2 |
POU class 2 homeobox 2 |
| chr11_+_71938925 | 0.07 |
ENST00000538751.1 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
| chr1_-_158301312 | 0.06 |
ENST00000368168.3 |
CD1B |
CD1b molecule |
| chr1_-_161519579 | 0.06 |
ENST00000426740.1 |
FCGR3A |
Fc fragment of IgG, low affinity IIIa, receptor (CD16a) |
| chr1_-_8000872 | 0.06 |
ENST00000377507.3 |
TNFRSF9 |
tumor necrosis factor receptor superfamily, member 9 |
| chr7_+_27282319 | 0.06 |
ENST00000222761.3 |
EVX1 |
even-skipped homeobox 1 |
| chr3_-_195538760 | 0.06 |
ENST00000475231.1 |
MUC4 |
mucin 4, cell surface associated |
| chr2_-_220436248 | 0.06 |
ENST00000265318.4 |
OBSL1 |
obscurin-like 1 |
| chr3_-_195538728 | 0.06 |
ENST00000349607.4 ENST00000346145.4 |
MUC4 |
mucin 4, cell surface associated |
| chr5_+_31193847 | 0.05 |
ENST00000514738.1 ENST00000265071.2 |
CDH6 |
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr17_-_19648916 | 0.05 |
ENST00000444455.1 ENST00000439102.2 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
| chr11_+_61717535 | 0.05 |
ENST00000534553.1 ENST00000301774.9 |
BEST1 |
bestrophin 1 |
| chr12_-_81331697 | 0.05 |
ENST00000552864.1 |
LIN7A |
lin-7 homolog A (C. elegans) |
| chrX_+_119495934 | 0.05 |
ENST00000218008.3 ENST00000361319.3 ENST00000539306.1 |
ATP1B4 |
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chr12_-_96390063 | 0.05 |
ENST00000541929.1 |
HAL |
histidine ammonia-lyase |
| chr12_-_48164812 | 0.05 |
ENST00000549151.1 ENST00000548919.1 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chrX_+_1387693 | 0.04 |
ENST00000381529.3 ENST00000432318.2 ENST00000361536.3 ENST00000501036.2 ENST00000381524.3 ENST00000412290.1 |
CSF2RA |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
| chr7_-_148823387 | 0.04 |
ENST00000483014.1 ENST00000378061.2 |
ZNF425 |
zinc finger protein 425 |
| chr3_-_58572760 | 0.04 |
ENST00000447756.2 |
FAM107A |
family with sequence similarity 107, member A |
| chr11_-_77791156 | 0.04 |
ENST00000281031.4 |
NDUFC2 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa |
| chr10_-_120514720 | 0.04 |
ENST00000369151.3 ENST00000340214.4 |
CACUL1 |
CDK2-associated, cullin domain 1 |
| chr12_-_96390108 | 0.04 |
ENST00000538703.1 ENST00000261208.3 |
HAL |
histidine ammonia-lyase |
| chr1_+_89829610 | 0.03 |
ENST00000370456.4 ENST00000535065.1 |
GBP6 |
guanylate binding protein family, member 6 |
| chr16_+_4674787 | 0.03 |
ENST00000262370.7 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr15_+_28623784 | 0.03 |
ENST00000526619.2 ENST00000337838.7 ENST00000532622.2 |
GOLGA8F |
golgin A8 family, member F |
| chr1_-_154580616 | 0.03 |
ENST00000368474.4 |
ADAR |
adenosine deaminase, RNA-specific |
| chr4_-_168155700 | 0.02 |
ENST00000357545.4 ENST00000512648.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr4_+_74718906 | 0.02 |
ENST00000226524.3 |
PF4V1 |
platelet factor 4 variant 1 |
| chr1_-_228603694 | 0.02 |
ENST00000366697.2 |
TRIM17 |
tripartite motif containing 17 |
| chr11_-_4414880 | 0.02 |
ENST00000254436.7 ENST00000543625.1 |
TRIM21 |
tripartite motif containing 21 |
| chr10_+_82009466 | 0.02 |
ENST00000356374.4 |
AL359195.1 |
Uncharacterized protein; cDNA FLJ46261 fis, clone TESTI4025062 |
| chr12_-_81331460 | 0.01 |
ENST00000549417.1 |
LIN7A |
lin-7 homolog A (C. elegans) |
| chr12_-_11548496 | 0.01 |
ENST00000389362.4 ENST00000565533.1 ENST00000546254.1 |
PRB2 PRB1 |
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr20_+_51588873 | 0.01 |
ENST00000371497.5 |
TSHZ2 |
teashirt zinc finger homeobox 2 |
| chr11_-_62783276 | 0.01 |
ENST00000535878.1 ENST00000545207.1 |
SLC22A8 |
solute carrier family 22 (organic anion transporter), member 8 |
| chr15_-_53082178 | 0.00 |
ENST00000305901.5 |
ONECUT1 |
one cut homeobox 1 |
| chr1_+_209929494 | 0.00 |
ENST00000367026.3 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
| chr17_+_73089382 | 0.00 |
ENST00000538213.2 ENST00000584118.1 |
SLC16A5 |
solute carrier family 16 (monocarboxylate transporter), member 5 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 2.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.3 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.8 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 2.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.2 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.3 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 2.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.5 | 1.5 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.3 | 0.8 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.2 | 0.5 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.2 | 2.0 | GO:0090240 | follicle-stimulating hormone signaling pathway(GO:0042699) positive regulation of histone H4 acetylation(GO:0090240) |
| 0.2 | 0.8 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 2.4 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.3 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.6 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.7 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 | 0.4 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.5 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.3 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.1 | 0.5 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.3 | GO:1904327 | maintenance of unfolded protein(GO:0036506) protein localization to cytosolic proteasome complex(GO:1904327) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.1 | 1.7 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.2 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.4 | GO:1902741 | interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.1 | 0.3 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.2 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.2 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.1 | 0.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.5 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.9 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.1 | GO:0021779 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0051946 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.3 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 1.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 1.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of B cell differentiation(GO:0045578) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.6 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.4 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.3 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.0 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.6 | GO:0008542 | visual learning(GO:0008542) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.5 | 1.5 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.3 | 0.8 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.2 | 0.9 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 3.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.1 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.7 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.3 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 0.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.5 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.2 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.1 | GO:0050135 | NAD+ nucleosidase activity(GO:0003953) NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.0 | 1.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 1.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.6 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 1.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 2.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 2.0 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.0 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |