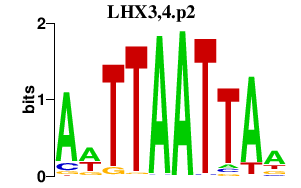
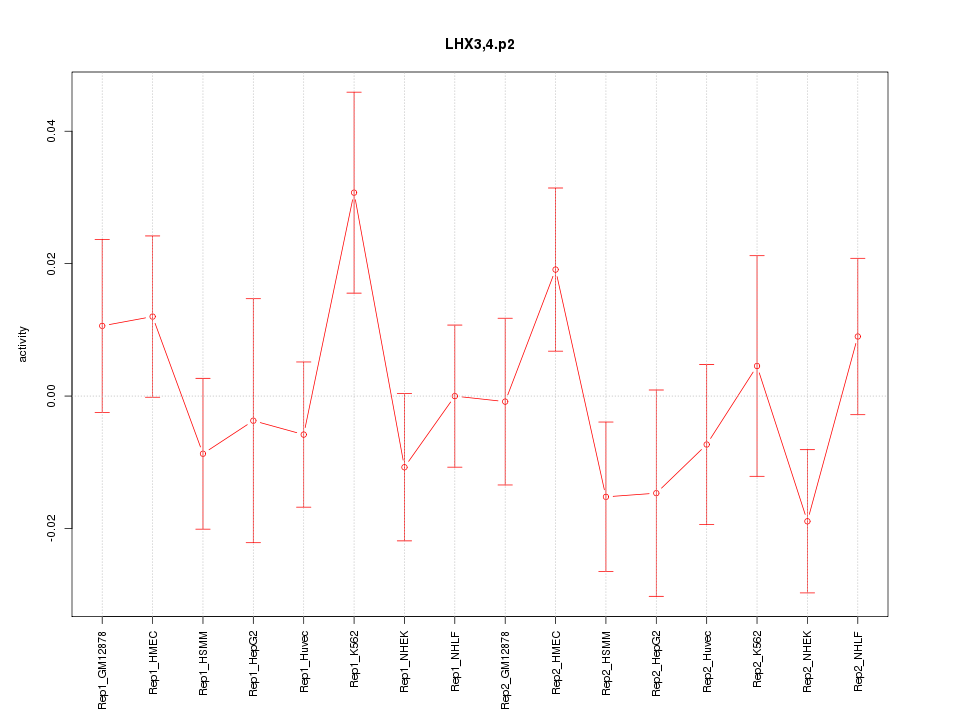
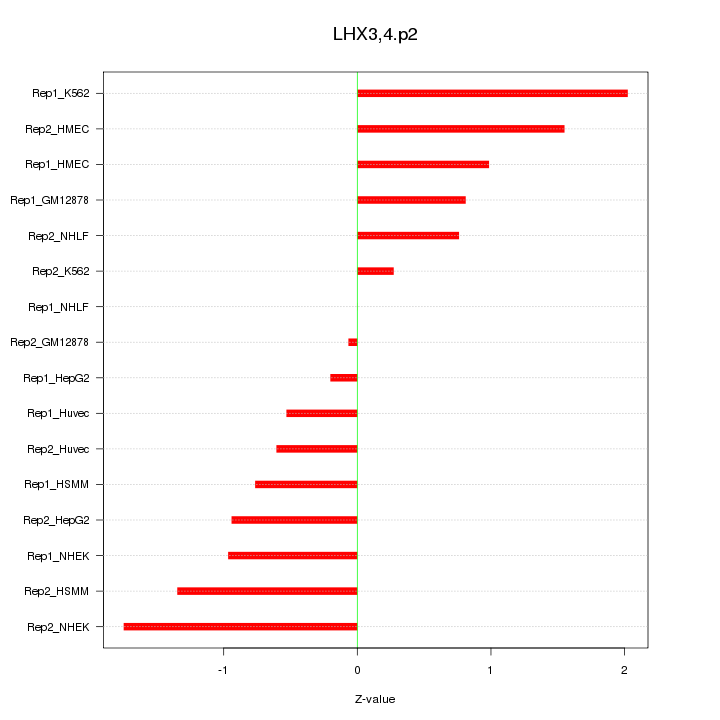
|
chr19_+_60962318
|
1.625
|
NM_001145014
|
RFPL4A
|
ret finger protein-like 4A
|
|
chr3_-_69254226
|
1.251
|
NM_198271
|
LMOD3
|
leiomodin 3 (fetal)
|
|
chr5_+_6799100
|
1.209
|
NM_001171806
|
PAPD7
|
PAP associated domain containing 7
|
|
chr8_-_131483395
|
1.025
|
NM_018482
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1
|
|
chr15_+_32048380
|
1.024
|
NM_012125
|
CHRM5
|
cholinergic receptor, muscarinic 5
|
|
chr6_+_73133152
|
0.980
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr15_+_100175888
|
0.976
|
NM_001001674
|
OR4F15
|
olfactory receptor, family 4, subfamily F, member 15
|
|
chr8_+_124032830
|
0.965
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr12_-_7547552
|
0.881
|
NM_004244
NM_203416
|
CD163
|
CD163 molecule
|
|
chr6_+_155579788
|
0.867
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr12_-_41269679
|
0.839
|
NM_153026
|
PRICKLE1
|
prickle homolog 1 (Drosophila)
|
|
chr5_-_16795336
|
0.796
|
|
MYO10
|
myosin X
|
|
chr1_-_93919789
|
0.790
|
NM_003567
|
BCAR3
|
breast cancer anti-estrogen resistance 3
|
|
chr7_+_129771865
|
0.783
|
NM_001127441
NM_001127442
NM_080385
|
CPA5
|
carboxypeptidase A5
|
|
chr1_+_156991133
|
0.778
|
NM_001005184
|
OR6K6
|
olfactory receptor, family 6, subfamily K, member 6
|
|
chr8_+_17440664
|
0.777
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chrX_+_7147471
|
0.740
|
NM_000351
|
STS
|
steroid sulfatase (microsomal), isozyme S
|
|
chr1_+_242281199
|
0.733
|
|
ZNF238
|
zinc finger protein 238
|
|
chr7_+_128802719
|
0.728
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr6_-_25679197
|
0.728
|
|
|
|
|
chr2_-_99238001
|
0.719
|
NM_175735
|
LYG2
|
lysozyme G-like 2
|
|
chr9_-_111230926
|
0.713
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr1_-_150398327
|
0.703
|
NM_001122965
|
RPTN
|
repetin
|
|
chr16_-_70168449
|
0.700
|
NM_000353
|
TAT
|
tyrosine aminotransferase
|
|
chr5_-_88155360
|
0.678
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr20_-_49747397
|
0.656
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr1_+_242281175
|
0.655
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chrX_-_55074125
|
0.631
|
NM_000032
NM_001037967
NM_001037968
|
ALAS2
|
aminolevulinate, delta-, synthase 2
|
|
chr1_-_159191184
|
0.623
|
NM_080878
|
ITLN2
|
intelectin 2
|
|
chr6_-_50045296
|
0.619
|
NM_001037729
|
DEFB113
|
defensin, beta 113
|
|
chr6_+_29663661
|
0.619
|
NM_007160
|
OR2H2
|
olfactory receptor, family 2, subfamily H, member 2
|
|
chr3_+_196933422
|
0.615
|
NM_152673
|
MUC20
|
mucin 20, cell surface associated
|
|
chr7_-_27133163
|
0.605
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr11_-_111636792
|
0.580
|
NM_001145024
|
C11orf34
|
chromosome 11 open reading frame 34
|
|
chr9_+_674193
|
0.578
|
|
|
|
|
chr4_-_170314382
|
0.577
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr10_+_685887
|
0.570
|
|
C10orf108
|
chromosome 10 open reading frame 108
|
|
chr1_+_242283129
|
0.568
|
NM_006352
|
ZNF238
|
zinc finger protein 238
|
|
chr3_+_46386636
|
0.567
|
NM_000579
NM_001100168
|
CCR5
|
chemokine (C-C motif) receptor 5
|
|
chr1_-_195818927
|
0.563
|
|
DENND1B
|
DENN/MADD domain containing 1B
|
|
chr14_+_69307914
|
0.561
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr7_-_86433139
|
0.548
|
NM_152748
|
KIAA1324L
|
KIAA1324-like
|
|
chrX_-_110542040
|
0.543
|
NM_178151
NM_001195553
NM_178152
NM_178153
|
DCX
|
doublecortin
|
|
chr11_+_33306652
|
0.539
|
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr5_-_137502975
|
0.536
|
NM_003551
|
NME5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr22_+_21817512
|
0.528
|
NM_004914
|
RAB36
|
RAB36, member RAS oncogene family
|
|
chr11_-_114593838
|
0.510
|
|
CADM1
|
cell adhesion molecule 1
|
|
chr21_-_34820916
|
0.502
|
NM_203418
|
RCAN1
|
regulator of calcineurin 1
|
|
chrX_-_43717864
|
0.498
|
NM_000266
|
NDP
|
Norrie disease (pseudoglioma)
|
|
chr2_-_224574860
|
0.494
|
|
SERPINE2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2
|
|
chr2_-_166638376
|
0.494
|
NM_001165963
NM_001165964
NM_006920
|
SCN1A
|
sodium channel, voltage-gated, type I, alpha subunit
|
|
chr10_+_21924265
|
0.491
|
|
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10
|
|
chr6_+_161043214
|
0.489
|
NM_000301
NM_001168338
|
PLG
|
plasminogen
|
|
chr8_+_22900863
|
0.477
|
NM_001160036
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr9_+_27099138
|
0.472
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr8_-_7207899
|
0.472
|
NM_001164457
|
ZNF705G
|
zinc finger protein 705G
|
|
chr3_+_46628928
|
0.469
|
NM_001195442
|
LOC100132146
|
hypothetical LOC100132146
|
|
chr1_-_203592538
|
0.464
|
NM_018203
|
KLHDC8A
|
kelch domain containing 8A
|
|
chr6_-_2580296
|
0.464
|
NM_152554
|
C6orf195
|
chromosome 6 open reading frame 195
|
|
chr2_+_178893216
|
0.463
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr20_+_56919233
|
0.463
|
|
GNAS
|
GNAS complex locus
|
|
chr8_-_7710547
|
0.462
|
NM_001037668
NM_001040705
|
DEFB107A
DEFB107B
|
defensin, beta 107A
defensin, beta 107B
|
|
chrX_-_15242589
|
0.461
|
NM_001012428
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chr17_-_36347197
|
0.460
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr10_-_48426868
|
0.458
|
NM_001042357
NM_001042389
|
PTPN20B
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20B
protein tyrosine phosphatase, non-receptor type 20A
|
|
chr3_+_152934393
|
0.452
|
NM_207365
|
AADACL2
|
arylacetamide deacetylase-like 2
|
|
chr7_-_122627235
|
0.450
|
NM_022444
|
SLC13A1
|
solute carrier family 13 (sodium/sulfate symporters), member 1
|
|
chr1_+_42773146
|
0.444
|
NM_001080850
|
CCDC30
|
coiled-coil domain containing 30
|
|
chr17_-_36877002
|
0.434
|
NM_002278
|
KRT32
|
keratin 32
|
|
chr11_+_124294355
|
0.428
|
NM_001037558
|
HEPN1
|
hepatocellular carcinoma, down-regulated 1
|
|
chr4_-_68511738
|
0.417
|
NM_001114387
NM_182606
|
TMPRSS11A
|
transmembrane protease, serine 11A
|
|
chr6_-_31196174
|
0.416
|
NM_001264
|
CDSN
|
corneodesmosin
|
|
chr7_+_147918589
|
0.414
|
NM_145304
|
C7orf33
|
chromosome 7 open reading frame 33
|
|
chr11_-_5819702
|
0.412
|
NM_001005167
|
OR52E6
|
olfactory receptor, family 52, subfamily E, member 6
|
|
chr13_+_52500830
|
0.409
|
NM_006418
|
OLFM4
|
olfactomedin 4
|
|
chr4_-_26100986
|
0.402
|
NM_000730
|
CCKAR
|
cholecystokinin A receptor
|
|
chr7_-_38279757
|
0.400
|
NM_001003799
NM_001003806
|
TARP
TRGV9
|
TCR gamma alternate reading frame protein
T cell receptor gamma variable 9
|
|
chr9_+_129066576
|
0.399
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr18_+_28023984
|
0.397
|
NM_005925
|
MEP1B
|
meprin A, beta
|
|
chr11_+_89403921
|
0.396
|
NM_001195234
|
TRIM49
TRIM49L2
|
tripartite motif containing 49
tripartite motif containing 49-like 2
|
|
chr6_+_136214495
|
0.394
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr5_+_36642424
|
0.394
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr20_-_29441946
|
0.394
|
NM_153289
NM_153323
NM_173460
|
DEFB119
|
defensin, beta 119
|
|
chr5_-_111121691
|
0.394
|
NM_001142477
NM_001142476
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr10_-_33264491
|
0.390
|
NM_033668
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr4_+_169789327
|
0.389
|
NM_001166109
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr6_-_152681171
|
0.388
|
NM_015293
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr9_-_14076901
|
0.388
|
|
NFIB
|
nuclear factor I/B
|
|
chr5_+_36642213
|
0.387
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr1_-_67039518
|
0.387
|
NM_005478
|
INSL5
|
insulin-like 5
|
|
chr8_+_7340777
|
0.386
|
NM_001037668
NM_001040705
|
DEFB107A
DEFB107B
|
defensin, beta 107A
defensin, beta 107B
|
|
chr13_-_109600709
|
0.385
|
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr2_-_87102453
|
0.384
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr16_-_71650034
|
0.384
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr5_-_160044844
|
0.383
|
|
ATP10B
|
ATPase, class V, type 10B
|
|
chr6_+_144654565
|
0.382
|
NM_007124
|
UTRN
|
utrophin
|
|
chr3_+_11269384
|
0.381
|
NM_000861
|
HRH1
|
histamine receptor H1
|
|
chr15_+_22832198
|
0.381
|
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr15_-_31147376
|
0.380
|
NM_001103184
|
FMN1
|
formin 1
|
|
chr16_+_2749934
|
0.380
|
|
|
|
|
chr6_-_29635680
|
0.379
|
NM_006398
|
UBD
|
ubiquitin D
|
|
chr5_-_37220702
|
0.378
|
|
C5orf42
|
chromosome 5 open reading frame 42
|
|
chr21_-_30460841
|
0.376
|
NM_012131
|
CLDN17
|
claudin 17
|
|
chr6_-_29120930
|
0.375
|
NM_030903
|
OR2W1
|
olfactory receptor, family 2, subfamily W, member 1
|
|
chrX_+_135558001
|
0.373
|
NM_000074
|
CD40LG
|
CD40 ligand
|
|
chr5_+_127012611
|
0.373
|
NM_001048252
|
CTXN3
|
cortexin 3
|
|
chr12_+_100394787
|
0.370
|
NM_152323
|
SPIC
|
Spi-C transcription factor (Spi-1/PU.1 related)
|
|
chr10_+_63478974
|
0.367
|
|
ARID5B
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr2_-_175170674
|
0.364
|
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chrX_-_44945025
|
0.359
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chr1_+_166516901
|
0.353
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr4_+_187227302
|
0.351
|
NM_003265
|
TLR3
|
toll-like receptor 3
|
|
chr16_-_3783475
|
0.350
|
|
CREBBP
|
CREB binding protein
|
|
chr10_+_24795446
|
0.347
|
|
KIAA1217
|
KIAA1217
|
|
chr2_+_135312655
|
0.347
|
NM_138326
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase
|
|
chr2_-_56266408
|
0.346
|
|
LOC100129434
|
hypothetical LOC100129434
|
|
chr3_+_153074118
|
0.343
|
NM_033050
|
SUCNR1
|
succinate receptor 1
|
|
chr1_+_200775864
|
0.342
|
NM_001197131
|
PPP1R12B
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B
|
|
chr11_+_89032112
|
0.340
|
NM_153696
|
FOLH1B
|
folate hydrolase 1B
|
|
chr7_+_111908143
|
0.338
|
NM_001134468
NM_182597
|
C7orf53
|
chromosome 7 open reading frame 53
|
|
chr3_-_127810087
|
0.336
|
NM_001039783
|
TXNRD3IT1
|
thioredoxin reductase 3 intronic transcript 1
|
|
chr11_-_117628211
|
0.334
|
NM_198275
|
MPZL3
|
myelin protein zero-like 3
|
|
chr19_-_40995564
|
0.333
|
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr5_+_145296313
|
0.332
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr11_-_18913121
|
0.329
|
NM_147199
|
MRGPRX1
|
MAS-related GPR, member X1
|
|
chr4_-_54119102
|
0.329
|
|
LNX1
|
ligand of numb-protein X 1
|
|
chr3_-_11598829
|
0.325
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chrX_-_138875342
|
0.323
|
NM_001013403
|
CXorf66
|
chromosome X open reading frame 66
|
|
chr1_-_163680941
|
0.323
|
NM_006917
|
RXRG
|
retinoid X receptor, gamma
|
|
chr3_-_15538164
|
0.320
|
NM_005677
NM_080539
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr12_+_54172413
|
0.320
|
NM_001005519
|
OR6C68
|
olfactory receptor, family 6, subfamily C, member 68
|
|
chr5_+_95092384
|
0.318
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr6_+_152053323
|
0.317
|
NM_001122742
|
ESR1
|
estrogen receptor 1
|
|
chr15_+_94670160
|
0.316
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr13_+_97856148
|
0.315
|
|
|
|
|
chr6_-_66346417
|
0.314
|
NM_198283
|
EYS
|
eyes shut homolog (Drosophila)
|
|
chr4_+_166350620
|
0.311
|
NM_001161521
NM_001161522
|
KLHL2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr19_-_40995606
|
0.310
|
|
PRODH2
|
proline dehydrogenase (oxidase) 2
|
|
chr19_+_21116666
|
0.309
|
NM_133473
|
ZNF431
|
zinc finger protein 431
|
|
chr12_-_52281050
|
0.308
|
NM_001130059
|
ATF7
|
activating transcription factor 7
|
|
chrX_-_18600108
|
0.308
|
NM_000330
|
RS1
|
retinoschisin 1
|
|
chr15_+_55298908
|
0.307
|
NM_207040
|
TCF12
|
transcription factor 12
|
|
chrX_-_62697473
|
0.306
|
|
LOC92249
|
hypothetical LOC92249
|
|
chr11_+_59979857
|
0.306
|
NM_021950
NM_152866
|
MS4A1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr11_+_61794205
|
0.306
|
NM_002411
|
SCGB2A2
|
secretoglobin, family 2A, member 2
|
|
chr17_-_36346240
|
0.305
|
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr1_-_157014048
|
0.304
|
NM_001005278
|
OR6N2
|
olfactory receptor, family 6, subfamily N, member 2
|
|
chr6_-_50097606
|
0.303
|
NM_001037497
NM_001037728
|
DEFB110
|
defensin, beta 110 locus
|
|
chr3_+_45902999
|
0.303
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chr14_-_105997732
|
0.302
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr10_+_24795465
|
0.301
|
|
KIAA1217
|
KIAA1217
|
|
chr21_-_30510113
|
0.300
|
NM_199328
|
CLDN8
|
claudin 8
|
|
chr18_+_59767567
|
0.299
|
NM_001123366
|
HMSD
|
histocompatibility (minor) serpin domain containing
|
|
chr1_-_152845399
|
0.298
|
NM_001193495
|
ADAR
|
adenosine deaminase, RNA-specific
|
|
chr5_-_124108672
|
0.298
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr19_-_56612653
|
0.295
|
NM_001171156
NM_001171157
NM_001171158
NM_001171159
NM_001171160
NM_001171161
NM_033130
|
SIGLEC10
|
sialic acid binding Ig-like lectin 10
|
|
chr2_+_158559936
|
0.291
|
NM_001135098
|
UPP2
|
uridine phosphorylase 2
|
|
chr3_+_115099007
|
0.286
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr7_+_137795387
|
0.286
|
NM_003852
NM_015905
|
TRIM24
|
tripartite motif containing 24
|
|
chr3_+_150675117
|
0.279
|
NM_004617
|
TM4SF4
|
transmembrane 4 L six family member 4
|
|
chr4_+_123311207
|
0.276
|
NM_015312
|
KIAA1109
|
KIAA1109
|
|
chrX_-_138552517
|
0.276
|
NM_001171877
NM_001171878
NM_001171879
NM_005369
|
MCF2
|
MCF.2 cell line derived transforming sequence
|
|
chr4_+_3341521
|
0.275
|
NM_198227
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr8_+_22266256
|
0.272
|
|
PIWIL2
|
piwi-like 2 (Drosophila)
|
|
chr11_-_89181390
|
0.271
|
NM_020358
|
TRIM49
|
tripartite motif containing 49
|
|
chr5_+_179065282
|
0.271
|
|
CANX
|
calnexin
|
|
chr9_+_117955890
|
0.269
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr15_-_64108718
|
0.269
|
|
|
|
|
chr9_-_15240113
|
0.268
|
NM_001168342
|
TTC39B
|
tetratricopeptide repeat domain 39B
|
|
chr4_-_54119159
|
0.265
|
NM_032622
|
LNX1
|
ligand of numb-protein X 1
|
|
chr1_+_119759076
|
0.265
|
NM_001166120
|
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2
|
|
chrX_+_50044279
|
0.265
|
NM_033031
NM_033670
|
CCNB3
|
cyclin B3
|
|
chr11_-_47472648
|
0.264
|
NM_198700
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr13_+_108046500
|
0.264
|
NM_015011
|
MYO16
|
myosin XVI
|
|
chr18_+_27210737
|
0.264
|
NM_001134453
NM_177986
|
DSG4
|
desmoglein 4
|
|
chr21_-_35174742
|
0.263
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_+_111634996
|
0.262
|
NM_021797
NM_201653
|
CHIA
|
chitinase, acidic
|
|
chr6_-_38671741
|
0.260
|
NM_001172418
NM_152733
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr20_+_56455866
|
0.260
|
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chrX_+_107174855
|
0.259
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr11_-_85115105
|
0.258
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr9_-_4142182
|
0.256
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr5_+_71538456
|
0.255
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr3_+_171022403
|
0.255
|
NM_001080460
|
LRRIQ4
|
leucine-rich repeats and IQ motif containing 4
|
|
chrX_+_55666557
|
0.253
|
NM_198451
|
FOXR2
|
forkhead box R2
|
|
chr1_-_155336223
|
0.253
|
NM_001004341
|
ETV3L
|
ets variant 3-like
|
|
chr2_-_206790871
|
0.252
|
NM_001098199
NM_005279
|
GPR1
|
G protein-coupled receptor 1
|
|
chr9_+_123369219
|
0.251
|
NM_032552
|
DAB2IP
|
DAB2 interacting protein
|
|
chr2_+_216695062
|
0.247
|
|
XRCC5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining)
|
|
chr7_-_139408808
|
0.246
|
|
PARP12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr7_+_18296569
|
0.245
|
|
HDAC9
|
histone deacetylase 9
|
|
chrX_+_70682204
|
0.243
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr4_+_111616630
|
0.242
|
NM_001977
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A)
|
|
chr12_-_10870134
|
0.237
|
NM_023921
|
TAS2R10
|
taste receptor, type 2, member 10
|
|
chrX_+_55951280
|
0.236
|
|
|
|
|
chr1_+_13903936
|
0.233
|
NM_012231
NM_015866
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr6_-_24985432
|
0.233
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr8_-_62764839
|
0.232
|
NM_001164750
NM_001164751
NM_001164752
NM_001164753
NM_020164
NM_032467
NM_032468
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr16_+_3473476
|
0.232
|
|
NAT15
|
N-acetyltransferase 15 (GCN5-related, putative)
|
|
chr19_+_9157269
|
0.232
|
NM_175883
|
OR7D2
|
olfactory receptor, family 7, subfamily D, member 2
|