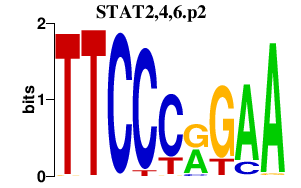
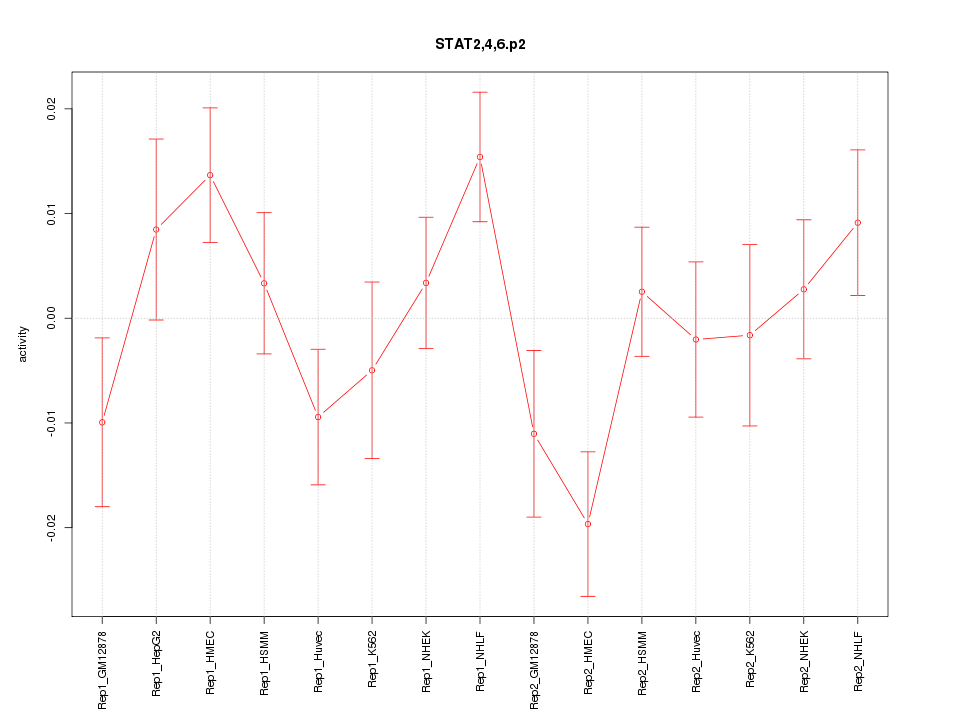
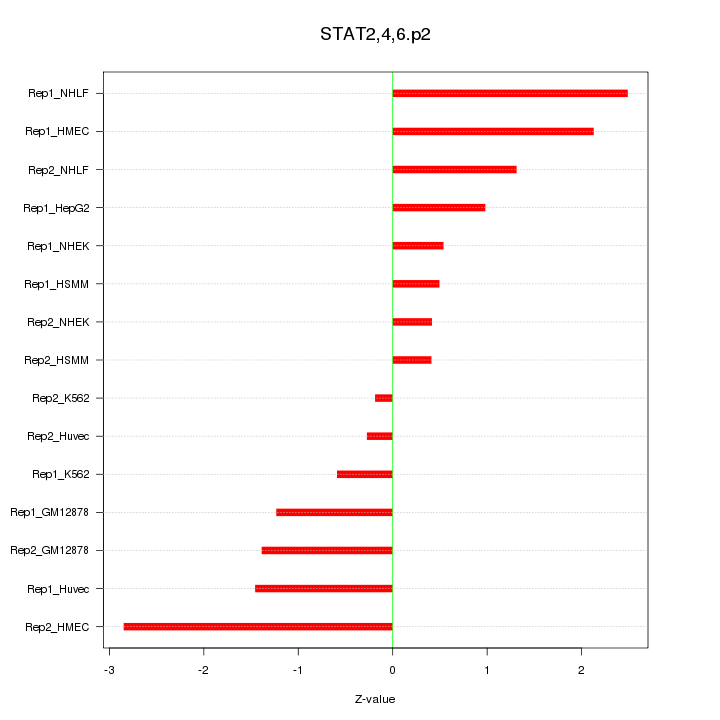
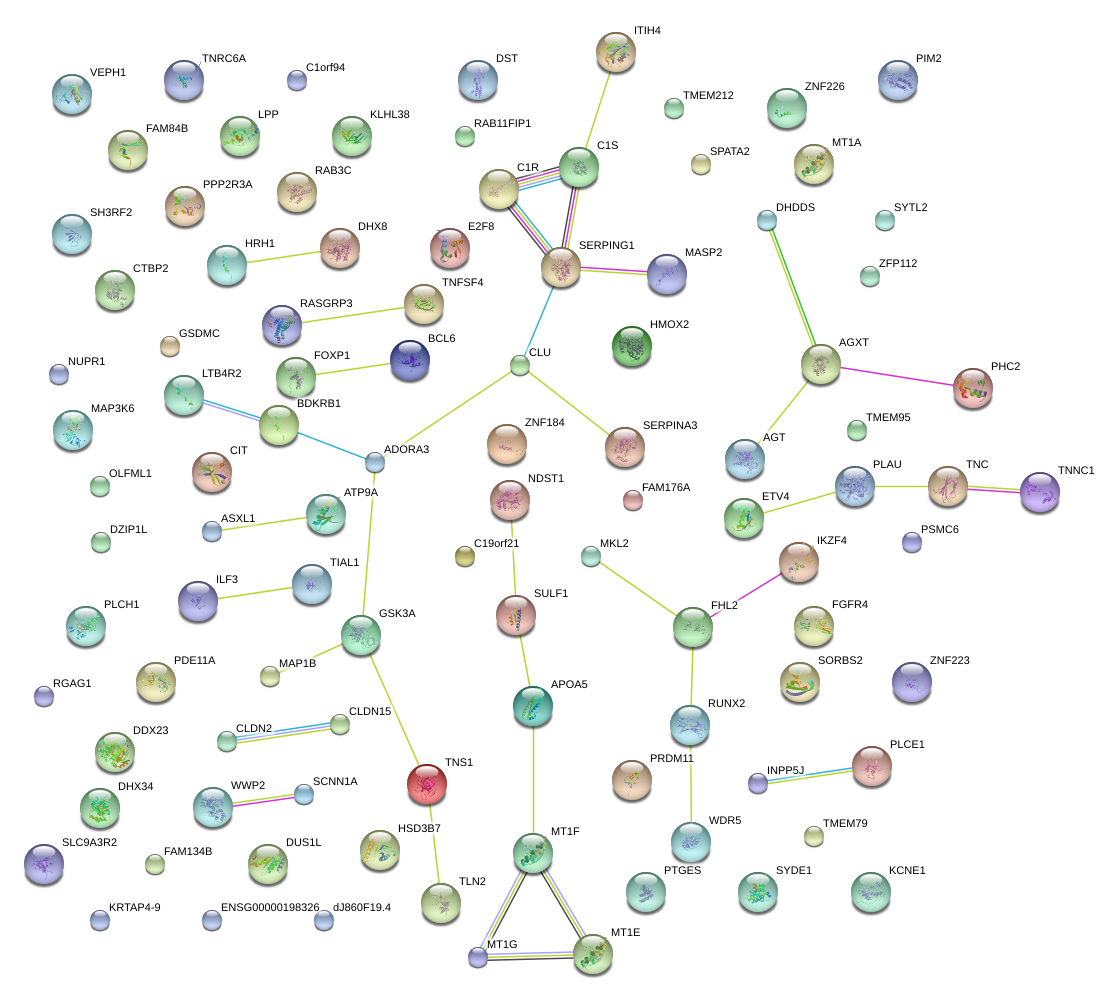
|
chr20_+_2743613
|
1.510
|
NM_080739
|
C20orf141
|
chromosome 20 open reading frame 141
|
|
chr3_+_189353502
|
1.459
|
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr10_+_75343352
|
1.302
|
|
PLAU
|
plasminogen activator, urokinase
|
|
chr19_-_49552638
|
1.281
|
NM_001083335
NM_013380
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr20_+_30409649
|
1.088
|
NM_001164603
NM_015338
|
ASXL1
|
additional sex combs like 1 (Drosophila)
|
|
chr10_+_95838801
|
1.071
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr20_-_48741481
|
0.994
|
|
|
|
|
chr19_+_49248003
|
0.977
|
NM_013361
|
ZNF223
|
zinc finger protein 223
|
|
chr5_+_71538456
|
0.951
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr15_+_60838053
|
0.941
|
|
TLN2
|
talin 2
|
|
chr14_+_95792299
|
0.926
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr12_+_7038240
|
0.901
|
NM_001734
NM_201442
|
C1S
|
complement component 1, s subcomponent
|
|
chr3_-_71376919
|
0.887
|
|
FOXP1
|
forkhead box P1
|
|
chr11_-_116168316
|
0.838
|
NM_001166598
NM_052968
|
APOA5
|
apolipoprotein A-V
|
|
chr14_+_94148466
|
0.771
|
NM_001085
|
SERPINA3
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3
|
|
chr12_+_54700955
|
0.749
|
NM_022465
|
IKZF4
|
IKAROS family zinc finger 4 (Eos)
|
|
chr11_+_57121602
|
0.717
|
NM_000062
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr3_-_52839727
|
0.716
|
NM_001166449
NM_002218
|
ITIH4
|
inter-alpha (globulin) inhibitor H4 (plasma Kallikrein-sensitive glycoprotein)
|
|
chr11_-_85115105
|
0.714
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr11_+_57122153
|
0.713
|
NM_001032295
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr3_-_156876798
|
0.698
|
NM_001130960
NM_001130961
|
PLCH1
|
phospholipase C, eta 1
|
|
chr12_-_7136147
|
0.698
|
NM_001733
|
C1R
|
complement component 1, r subcomponent
|
|
chrX_+_106030049
|
0.697
|
NM_001171092
|
CLDN2
|
claudin 2
|
|
chr1_-_27565923
|
0.686
|
NM_004672
|
MAP3K6
|
mitogen-activated protein kinase kinase kinase 6
|
|
chr9_+_135994734
|
0.686
|
NM_052821
|
WDR5
|
WD repeat domain 5
|
|
chr5_+_149867866
|
0.682
|
NM_001543
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr6_-_27548438
|
0.671
|
NM_007149
|
ZNF184
|
zinc finger protein 184
|
|
chr8_-_27525084
|
0.666
|
NM_001171138
|
CLU
|
clusterin
|
|
chr12_-_47532171
|
0.660
|
NM_004818
|
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23
|
|
chr2_-_218516950
|
0.654
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr12_-_7136189
|
0.652
|
|
C1R
|
complement component 1, r subcomponent
|
|
chr6_-_56615544
|
0.630
|
NM_001723
NM_015548
|
DST
|
dystonin
|
|
chr22_+_29848923
|
0.628
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr9_+_676640
|
0.617
|
|
|
|
|
chr5_+_176449156
|
0.616
|
NM_022963
|
FGFR4
|
fibroblast growth factor receptor 4
|
|
chrX_-_48661226
|
0.614
|
NM_006875
|
PIM2
|
pim-2 oncogene
|
|
chr16_+_14187965
|
0.572
|
|
MKL2
|
MKL/myocardin-like 2
|
|
chr19_+_702108
|
0.564
|
NM_173481
|
C19orf21
|
chromosome 19 open reading frame 21
|
|
chr5_+_145296313
|
0.560
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr17_+_36515166
|
0.536
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr3_+_137224266
|
0.530
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr3_-_139315011
|
0.529
|
NM_001170538
|
DZIP1L
|
DAZ interacting protein 1-like
|
|
chr6_-_56824672
|
0.528
|
|
DST
|
dystonin
|
|
chrX_+_109548940
|
0.523
|
NM_020769
|
RGAG1
|
retrotransposon gag domain containing 1
|
|
chr2_-_178645718
|
0.514
|
NM_016953
|
PDE11A
|
phosphodiesterase 11A
|
|
chr7_-_100667750
|
0.513
|
NM_014343
|
CLDN15
|
claudin 15
|
|
chr17_+_7199178
|
0.513
|
NM_198154
|
TMEM95
|
transmembrane protein 95
|
|
chr16_+_4485859
|
0.509
|
NM_001127205
|
HMOX2
|
heme oxygenase (decycling) 2
|
|
chr1_-_20682541
|
0.506
|
|
|
|
|
chr8_-_27524744
|
0.505
|
NM_203339
|
CLU
|
clusterin
|
|
chr9_-_116920232
|
0.505
|
NM_002160
|
TNC
|
tenascin C
|
|
chr9_+_22103542
|
0.502
|
|
CDKN2B-AS1
|
CDKN2B antisense RNA 1 (non-protein coding)
|
|
chr20_-_49747397
|
0.502
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr21_-_34749908
|
0.500
|
NM_001127670
|
KCNE1
|
potassium voltage-gated channel, Isk-related family, member 1
|
|
chr19_+_15079141
|
0.492
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr2_+_33554824
|
0.488
|
NM_001139488
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr14_+_52243645
|
0.487
|
NM_002806
|
PSMC6
|
proteasome (prosome, macropain) 26S subunit, ATPase, 6
|
|
chr8_-_130868315
|
0.487
|
NM_031415
|
GSDMC
|
gasdermin C
|
|
chr20_-_47963682
|
0.479
|
NM_001135773
|
SPATA2
|
spermatogenesis associated 2
|
|
chr1_-_228916958
|
0.478
|
NM_000029
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr17_-_38979178
|
0.471
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr1_+_154520693
|
0.469
|
NM_032323
|
TMEM79
|
transmembrane protein 79
|
|
chr10_-_121346517
|
0.469
|
NM_001033925
NM_003252
|
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1
|
|
chr8_+_70567581
|
0.466
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr19_+_54996613
|
0.464
|
|
|
|
|
chr8_-_37847095
|
0.461
|
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr1_-_11029882
|
0.460
|
|
MASP2
|
mannan-binding lectin serine peptidase 2
|
|
chr3_+_11153778
|
0.456
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr11_+_7463175
|
0.452
|
NM_198474
|
OLFML1
|
olfactomedin-like 1
|
|
chr1_-_228916559
|
0.450
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr8_-_124734346
|
0.448
|
NM_001081675
|
KLHL38
|
kelch-like 38 (Drosophila)
|
|
chr16_-_55259405
|
0.447
|
NM_005950
|
MT1G
|
metallothionein 1G
|
|
chr1_-_228916486
|
0.445
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr10_-_126706442
|
0.443
|
NM_022802
|
CTBP2
|
C-terminal binding protein 2
|
|
chr19_+_49361054
|
0.443
|
NM_001146220
NM_001032373
NM_001032374
NM_015919
NM_001032372
|
ZNF226
|
zinc finger protein 226
|
|
chr16_+_24696069
|
0.442
|
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr11_+_45072139
|
0.442
|
NM_020229
|
PRDM11
|
PR domain containing 11
|
|
chr16_+_30903963
|
0.440
|
NM_001142777
NM_001142778
NM_025193
|
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7
|
|
chr1_-_228916470
|
0.439
|
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr19_+_10642648
|
0.435
|
|
ILF3
|
interleukin enhancer binding factor 3, 90kDa
|
|
chr2_-_75650355
|
0.433
|
NM_032181
|
FAM176A
|
family with sequence similarity 176, member A
|
|
chr6_+_45404031
|
0.432
|
NM_001015051
NM_001024630
|
RUNX2
|
runt-related transcription factor 2
|
|
chr10_-_121346464
|
0.422
|
|
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1
|
|
chr1_-_111908085
|
0.422
|
NM_001081976
|
ADORA3
|
adenosine A3 receptor
|
|
chr2_-_105421661
|
0.418
|
NM_201557
|
FHL2
|
four and a half LIM domains 2
|
|
chr16_-_28457728
|
0.417
|
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr12_-_6356783
|
0.416
|
NM_001159575
|
SCNN1A
|
sodium channel, nonvoltage-gated 1 alpha
|
|
chr19_-_47438518
|
0.411
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr3_-_188935363
|
0.407
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr5_-_16561936
|
0.407
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr8_-_127639892
|
0.406
|
NM_174911
|
FAM84B
|
family with sequence similarity 84, member B
|
|
chr1_-_33613729
|
0.404
|
NM_198040
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr1_+_34405070
|
0.403
|
NM_032884
|
C1orf94
|
chromosome 1 open reading frame 94
|
|
chr1_-_171441234
|
0.403
|
|
TNFSF4
|
tumor necrosis factor (ligand) superfamily, member 4
|
|
chr16_+_68516401
|
0.400
|
NM_199424
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr3_+_173043832
|
0.399
|
NM_001164436
|
TMEM212
|
transmembrane protein 212
|
|
chr5_+_57914658
|
0.399
|
NM_138453
|
RAB3C
|
RAB3C, member RAS oncogene family
|
|
chr16_+_2025922
|
0.398
|
|
SLC9A3R2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr12_-_118725672
|
0.397
|
|
CIT
|
citron (rho-interacting, serine/threonine kinase 21)
|
|
chr1_-_11029737
|
0.394
|
NM_006610
NM_139208
|
MASP2
|
mannan-binding lectin serine peptidase 2
|
|
chr3_-_158700036
|
0.393
|
NM_024621
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr4_-_186934059
|
0.392
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr9_-_131555111
|
0.391
|
NM_004878
|
PTGES
|
prostaglandin E synthase
|
|
chr14_+_23849196
|
0.389
|
NM_001164692
NM_019839
|
LTB4R2
|
leukotriene B4 receptor 2
|
|
chr13_+_75092570
|
0.389
|
NM_005358
|
LMO7
|
LIM domain 7
|
|
chr11_-_11986704
|
0.388
|
NM_001018057
|
DKK3
|
dickkopf homolog 3 (Xenopus laevis)
|
|
chr9_-_90456824
|
0.388
|
NM_001100111
|
LOC286238
|
hypothetical protein LOC286238
|
|
chr22_+_38090120
|
0.387
|
NM_145738
|
SYNGR1
|
synaptogyrin 1
|
|
chr16_+_29378694
|
0.385
|
NM_001017390
NM_177552
|
SULT1A4
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 4
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3
|
|
chr6_+_31190505
|
0.383
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr3_-_102888090
|
0.381
|
NM_000986
|
RPL24
|
ribosomal protein L24
|
|
chr12_+_79362256
|
0.380
|
NM_001145026
|
PTPRQ
|
protein tyrosine phosphatase, receptor type, Q
|
|
chr7_+_129634935
|
0.379
|
NM_145268
|
C7orf45
|
chromosome 7 open reading frame 45
|
|
chr5_+_131437383
|
0.378
|
NM_000758
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage)
|
|
chr7_+_134114912
|
0.376
|
|
CALD1
|
caldesmon 1
|
|
chr7_-_133652042
|
0.374
|
|
SLC35B4
|
solute carrier family 35, member B4
|
|
chr16_-_15644465
|
0.369
|
NM_001184998
NM_001184999
NM_014647
|
KIAA0430
|
KIAA0430
|
|
chr1_+_154120170
|
0.369
|
|
SYT11
|
synaptotagmin XI
|
|
chr8_-_120720200
|
0.366
|
NM_001040092
NM_001130863
NM_006209
|
ENPP2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2
|
|
chr7_+_134114957
|
0.365
|
|
CALD1
|
caldesmon 1
|
|
chr8_+_70567634
|
0.363
|
|
SULF1
|
sulfatase 1
|
|
chr5_-_111340421
|
0.362
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr19_-_47438548
|
0.360
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr3_+_52803823
|
0.358
|
NM_002217
|
ITIH3
|
inter-alpha (globulin) inhibitor H3
|
|
chr16_+_15644624
|
0.357
|
NM_001143979
|
NDE1
|
nudE nuclear distribution gene E homolog 1 (A. nidulans)
|
|
chr9_+_124836666
|
0.356
|
NM_005294
|
GPR21
|
G protein-coupled receptor 21
|
|
chr3_-_48607596
|
0.356
|
NM_000094
|
COL7A1
|
collagen, type VII, alpha 1
|
|
chr14_+_54238581
|
0.356
|
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr19_-_47438563
|
0.356
|
NM_019884
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr14_-_22516271
|
0.356
|
NM_198086
|
JUB
|
jub, ajuba homolog (Xenopus laevis)
|
|
chr17_-_51155073
|
0.354
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr7_-_83116414
|
0.351
|
NM_012431
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr1_-_151788341
|
0.349
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr15_-_53996597
|
0.344
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr12_-_51200347
|
0.344
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr15_+_79304694
|
0.342
|
NM_172217
|
IL16
|
interleukin 16 (lymphocyte chemoattractant factor)
|
|
chr5_+_140454382
|
0.342
|
NM_018936
|
PCDHB2
|
protocadherin beta 2
|
|
chr2_-_180319013
|
0.341
|
NM_001113397
|
ZNF385B
|
zinc finger protein 385B
|
|
chr1_-_36589744
|
0.339
|
|
|
|
|
chr9_+_130722994
|
0.338
|
NM_001100876
NM_174933
|
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1
|
|
chr5_+_150008030
|
0.338
|
|
SYNPO
|
synaptopodin
|
|
chr5_-_140875591
|
0.332
|
|
DIAPH1
|
diaphanous homolog 1 (Drosophila)
|
|
chr7_-_149669622
|
0.330
|
NM_002889
|
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2
|
|
chr16_-_66535923
|
0.330
|
|
SLC12A4
|
solute carrier family 12 (potassium/chloride transporters), member 4
|
|
chr22_-_29866339
|
0.329
|
NM_015715
|
PLA2G3
|
phospholipase A2, group III
|
|
chr6_-_18372617
|
0.328
|
NM_001134709
NM_003472
|
DEK
|
DEK oncogene
|
|
chr3_+_186483408
|
0.328
|
|
LOC100505749
MAP3K13
|
hypothetical LOC100505749
mitogen-activated protein kinase kinase kinase 13
|
|
chr6_-_116973465
|
0.328
|
NM_001139444
|
BET3L
|
BET3 like (S. cerevisiae)
|
|
chr14_+_37746937
|
0.327
|
NM_001049
|
SSTR1
|
somatostatin receptor 1
|
|
chr16_-_28457825
|
0.326
|
NM_001042483
NM_012385
|
NUPR1
|
nuclear protein, transcriptional regulator, 1
|
|
chr7_-_14847479
|
0.322
|
NM_004080
NM_145695
|
DGKB
|
diacylglycerol kinase, beta 90kDa
|
|
chr4_-_87593306
|
0.316
|
NM_002753
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr11_-_8788765
|
0.316
|
NM_213618
|
ST5
|
suppression of tumorigenicity 5
|
|
chr11_-_62218759
|
0.314
|
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin)
|
|
chr9_+_116943917
|
0.313
|
NM_017418
|
DEC1
|
deleted in esophageal cancer 1
|
|
chr10_+_92621677
|
0.312
|
NM_001104546
NM_006413
|
RPP30
|
ribonuclease P/MRP 30kDa subunit
|
|
chr5_+_145807059
|
0.310
|
NM_001040006
NM_006706
|
TCERG1
|
transcription elongation regulator 1
|
|
chr3_-_64186151
|
0.308
|
NM_198859
|
PRICKLE2
|
prickle homolog 2 (Drosophila)
|
|
chr17_+_29606408
|
0.307
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr6_-_56824371
|
0.307
|
|
DST
|
dystonin
|
|
chr6_-_26393705
|
0.307
|
NM_003543
|
HIST1H4H
|
histone cluster 1, H4h
|
|
chr17_-_36528113
|
0.306
|
NM_033059
|
KRTAP4-11
|
keratin associated protein 4-11
|
|
chr12_-_68369322
|
0.305
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr5_+_178300799
|
0.304
|
NM_001178089
NM_001178090
NM_182594
|
ZNF454
|
zinc finger protein 454
|
|
chr6_-_42218692
|
0.299
|
NM_001164446
|
C6orf132
|
chromosome 6 open reading frame 132
|
|
chr19_-_54258935
|
0.297
|
NM_006179
|
NTF4
|
neurotrophin 4
|
|
chr10_-_116434364
|
0.297
|
NM_001003407
NM_001003408
|
ABLIM1
|
actin binding LIM protein 1
|
|
chr11_-_66431910
|
0.296
|
NM_022172
|
PC
|
pyruvate carboxylase
|
|
chr18_+_30427279
|
0.296
|
NM_001128175
NM_001198938
NM_001198939
NM_001198940
|
DTNA
|
dystrobrevin, alpha
|
|
chr14_-_50204771
|
0.295
|
NM_021818
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr3_+_189354167
|
0.295
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr22_+_19458378
|
0.294
|
NM_000185
|
SERPIND1
|
serpin peptidase inhibitor, clade D (heparin cofactor), member 1
|
|
chr14_-_22468393
|
0.293
|
|
PRMT5
|
protein arginine methyltransferase 5
|
|
chr4_-_40326603
|
0.292
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr4_+_122941921
|
0.285
|
NM_001034194
NM_005033
|
EXOSC9
|
exosome component 9
|
|
chr19_+_61742111
|
0.285
|
NM_020828
|
ZFP28
|
zinc finger protein 28 homolog (mouse)
|
|
chr12_+_52180971
|
0.285
|
NM_004178
NM_134323
|
TARBP2
|
TAR (HIV-1) RNA binding protein 2
|
|
chr12_+_50913220
|
0.284
|
NM_005556
|
KRT7
|
keratin 7
|
|
chr14_-_50204791
|
0.283
|
|
SAV1
|
salvador homolog 1 (Drosophila)
|
|
chr5_+_172415891
|
0.283
|
NM_001168393
NM_001168394
NM_153607
|
C5orf41
|
chromosome 5 open reading frame 41
|
|
chr14_-_24588916
|
0.282
|
NM_014178
|
STXBP6
|
syntaxin binding protein 6 (amisyn)
|
|
chr2_+_234624084
|
0.281
|
NM_006944
|
SPP2
|
secreted phosphoprotein 2, 24kDa
|
|
chr15_+_46411646
|
0.280
|
NM_001948
|
DUT
|
deoxyuridine triphosphatase
|
|
chr12_+_50913274
|
0.278
|
|
KRT7
|
keratin 7
|
|
chr19_-_47438583
|
0.277
|
|
GSK3A
|
glycogen synthase kinase 3 alpha
|
|
chr7_-_87774130
|
0.277
|
NM_024636
|
STEAP4
|
STEAP family member 4
|
|
chr3_+_116824840
|
0.274
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr14_+_37748031
|
0.272
|
|
SSTR1
|
somatostatin receptor 1
|
|
chr9_-_130526192
|
0.272
|
NM_032799
|
ZDHHC12
|
zinc finger, DHHC-type containing 12
|
|
chr6_+_36752244
|
0.270
|
NM_078467
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr7_+_21434097
|
0.269
|
NM_003112
|
SP4
|
Sp4 transcription factor
|
|
chr5_+_67558217
|
0.268
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr22_+_40286957
|
0.268
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chr5_-_138238877
|
0.267
|
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr5_+_149717489
|
0.266
|
|
TCOF1
|
Treacher Collins-Franceschetti syndrome 1
|
|
chr3_+_45042754
|
0.264
|
NM_003278
|
CLEC3B
|
C-type lectin domain family 3, member B
|
|
chr16_+_70646008
|
0.263
|
NM_001126102
NM_005143
|
HP
|
haptoglobin
|
|
chr22_-_35011703
|
0.262
|
|
MYH9
|
myosin, heavy chain 9, non-muscle
|
|
chr6_+_36754433
|
0.261
|
NM_000389
|
CDKN1A
|
cyclin-dependent kinase inhibitor 1A (p21, Cip1)
|
|
chr5_+_149717394
|
0.259
|
NM_000356
NM_001008657
NM_001135243
NM_001135244
NM_001135245
NM_001195141
|
TCOF1
|
Treacher Collins-Franceschetti syndrome 1
|