ISMARA results for ENCODE cell lines, H3K4me2 (Ernst 2011)
ISMARA - Integrated System for Motif Actitivity Response Analysis is a free online tool that recognizes most important transcription factors that are changing their activity in a set of samples.
All motifs sorted by activity significance
| Motif name | Z-value | Associated genes | Profile | Logo |
|---|---|---|---|---|
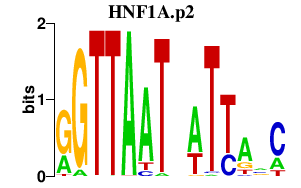
| HNF1A.p2 | 9.616 |
HNF1A
(HNF1, LFB1)
|

|
|
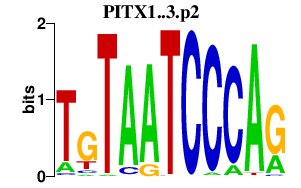
| PITX1..3.p2 | 9.583 |
PITX2
(IGDS, RS, Brx1, Otlx2, ARP1)
PITX3 PITX1 (PTX1, POTX) |
|
|
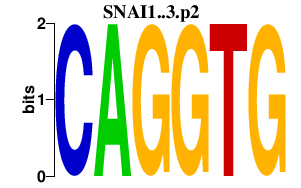
| SNAI1..3.p2 | 8.184 |
SNAI2
(SLUGH1)
SNAI1 (SNA, SLUGH2, SNAH) SNAI3 (SMUC, Zfp293) |
|
|
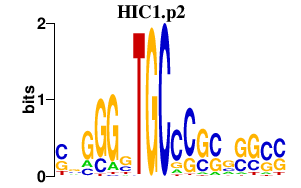
| HIC1.p2 | 7.893 |
HIC1
(ZBTB29)
|
|
|
| HNF4A_NR2F1,2.p2 | 7.784 |
NR2F1
(EAR-3, COUP-TFI, TCFCOUP1, SVP44)
NR2F2 (COUP-TFII, COUPTFB, SVP40) HNF4A (NR2A1, HNF4) |
|
|
| NRF1.p2 | 7.419 |
NRF1
(EWG, ALPHA-PAL)
|
|
|
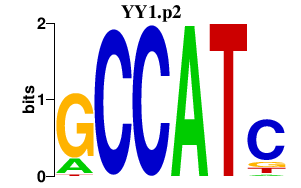
| YY1.p2 | 6.619 |
YY1
(NF-E1, DELTA, UCRBP, YIN-YANG-1, INO80S)
|
|
|
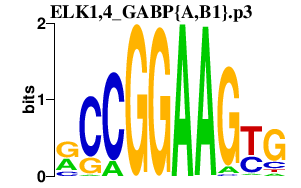
| ELK1,4_GABP{A,B1}.p3 | 5.725 |
GABPA
(E4TF1A, NFT2, NRF2, E4TF1-60, NRF2A)
GABPB1 (E4TF1-47, GABPB) ELK4 (SAP1) ELK1 |
|
|
| MTF1.p2 | 5.556 |
MTF1
|
|
|
| RUNX1..3.p2 | 5.272 |
RUNX1
(PEBP2A2, AMLCR1)
RUNX3 (AML2, PEBP2A3) RUNX2 (AML3, PEBP2A1, PEBP2aA1) |
|
|
| FOS_FOS{B,L1}_JUN{B,D}.p2 | 5.101 |
FOS
(c-fos, AP-1)
JUNB FOSL1 (fra-1) JUND (AP-1) FOSB (G0S3, GOSB, GOS3, AP-1, MGC42291, DKFZp686C0818) |

|

|
| HOX{A6,A7,B6,B7}.p2 | 4.884 |
HOXB6
HOXA7 HOXB7 HOXA6 |

|

|
| PRRX1,2.p2 | 4.879 |
PRRX2
(PRX2, PMX2)
PRRX1 (PHOX1) |
|

|
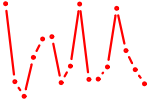
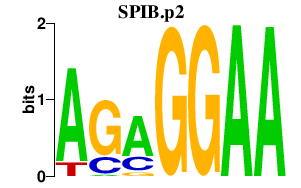
| SPIB.p2 | 4.623 |
SPIB
(SPI-B)
|
|
|
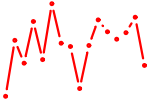
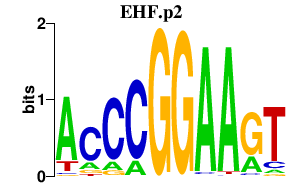
| EHF.p2 | 4.598 |
EHF
|
|
|
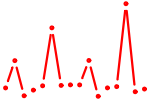
| GATA6.p2 | 4.543 |
GATA6
|
|
|
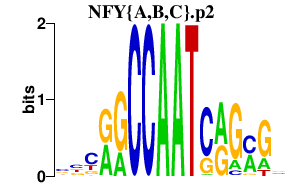
| NFY{A,B,C}.p2 | 4.386 |
NFYC
(CBF-C)
NFYB (CBF-A, HAP3) NFYA (HAP2, CBF-B) |
|
|
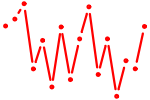
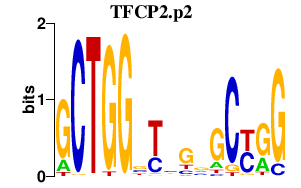
| TFCP2.p2 | 4.343 |
TFCP2
(CP2, LSF, LBP-1C, TFCP2C)
|
|
|
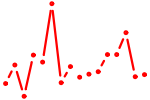
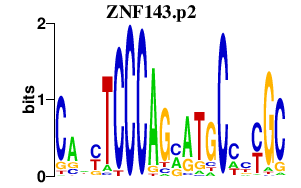
| ZNF143.p2 | 4.231 |
ZNF143
(SBF, pHZ-1, STAF)
|
|
|
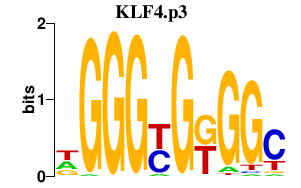
| KLF4.p3 | 4.228 |
KLF4
(EZF, GKLF)
|
|
|
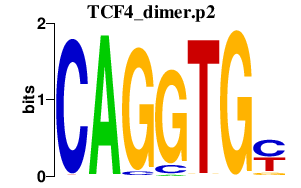
| TCF4_dimer.p2 | 3.999 |
TCF4
(E2-2, ITF2, PTHS, SEF2, SEF2-1, SEF2-1A, SEF2-1B, bHLHb19, MGC149723, MGC149724)
|
|
|
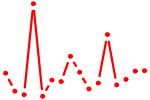
| MEF2{A,B,C,D}.p2 | 3.962 |
MEF2A
(RSRFC4, RSRFC9)
MEF2C MEF2D MEF2BNB-MEF2B (MEF2B, LOC729991-MEF2B) MEF2B (RSRFR2, FLJ32648) |
|
|
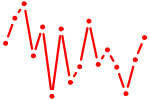
| TFAP2{A,C}.p2 | 3.910 |
TFAP2C
(AP2-GAMMA, ERF1, TFAP2G, hAP-2g)
TFAP2A (AP-2) |
|
|
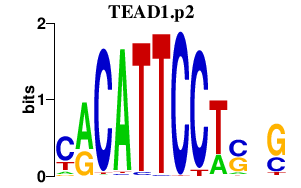
| TEAD1.p2 | 3.872 |
TEAD1
(TEF-1)
TEAD2 (ETF, TEF4, TEF-4, TEAD-2) TEAD3 (TEF5, TEAD5, TEF-5, DTEF-1, ETFR-1, TEAD-3) TEAD4 (TEF3, RTEF1, TEF-3, EFTR-2, TEFR-1, TCF13L1, hRTEF-1B) |
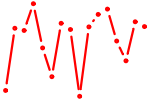
|
|
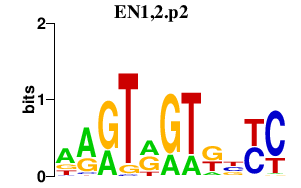
| EN1,2.p2 | 3.867 |
EN1
EN2 |

|
|
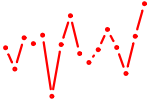
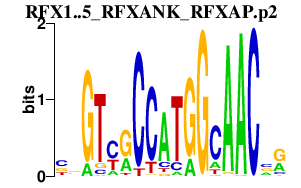
| RFX1..5_RFXANK_RFXAP.p2 | 3.765 |
RFX1
(EF-C)
RFXAP RFX5 RFXANK (BLS, RFX-B, ANKRA1, F14150_1, MGC138628) RFX4 RFX3 RFX2 (FLJ14226) |
|
|
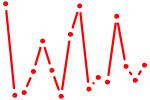
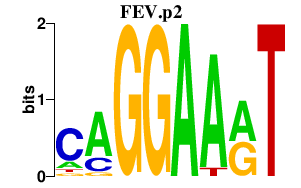
| FEV.p2 | 3.732 |
FEV
(PET-1, HSRNAFEV)
|
|
|
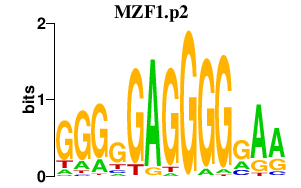
| MZF1.p2 | 3.708 |
MZF1
(MZF1B, ZSCAN6, MZF-1, Zfp98)
|

|
|
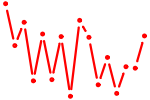
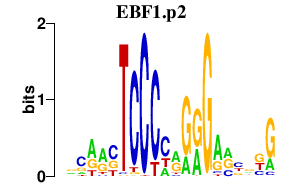
| EBF1.p2 | 3.574 |
EBF1
(OLF1)
|
|
|
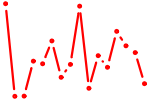
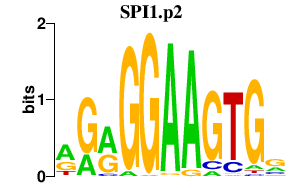
| SPI1.p2 | 3.456 |
SPI1
(PU.1, SPI-A, OF, SFPI1, SPI-1)
|
|
|
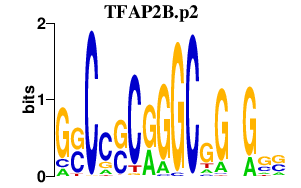
| TFAP2B.p2 | 3.420 |
TFAP2B
(AP2-B)
|
|
|
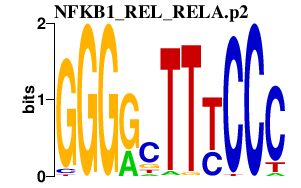
| NFKB1_REL_RELA.p2 | 3.360 |
RELA
(p65)
REL (I-Rel) NFKB1 (KBF1, p105) |
|
|
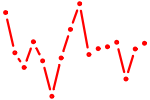
| IRF1,2,7.p3 | 3.349 |
IRF1
(MAR, IRF-1)
IRF2 (IRF-2, DKFZp686F0244) IRF7 (IRF7A, IRF-7H) |
|
|
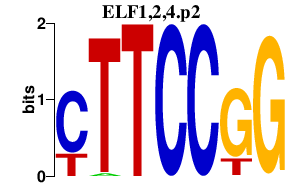
| ELF1,2,4.p2 | 3.292 |
ELF2
(EU32, NERF, NERF-2, NERF-1A, NERF-1B)
ELF1 ELF4 (MEF, ELFR) |
|
|
| FOXA2.p3 | 3.261 |
FOXA2
|
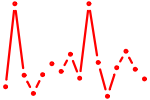
|

|
| ETS1,2.p2 | 3.213 |
ETS2
ETS1 (FLJ10768, ETS-1) |
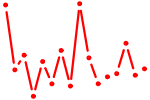
|
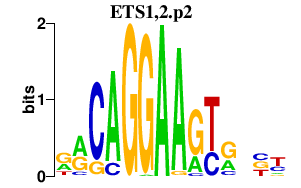
|
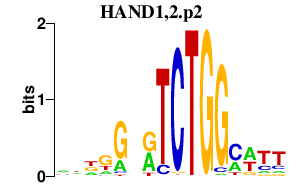
| HAND1,2.p2 | 3.203 |
HAND1
(eHand, Thing1, Hxt, bHLHa27)
HAND2 (dHand, Thing2, Hed, bHLHa26) |
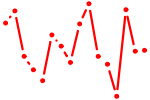
|
|
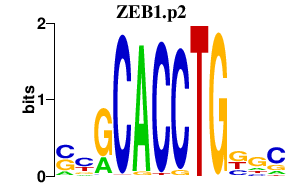
| ZEB1.p2 | 3.175 |
ZEB1
(BZP, ZEB, AREB6, NIL-2-A, Zfhep, Zfhx1a)
|
|
|
| MAFB.p2 | 3.159 |
MAFB
|
|
|
| NHLH1,2.p2 | 3.154 |
NHLH1
(NSCL, NSCL1, bHLHa35)
NHLH2 (NSCL2, bHLHa34) |

|
|
| KLF12.p2 | 3.094 |
KLF12
(AP-2rep, HSPC122, AP2REP)
|
|
|
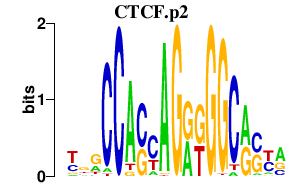
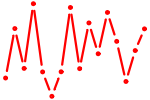
| CTCF.p2 | 3.031 |
CTCF
|

|
|
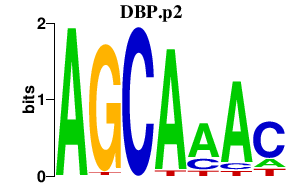
| DBP.p2 | 2.997 |
DBP
(DABP)
|
|
|
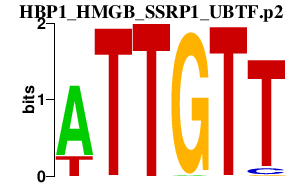
| HBP1_HMGB_SSRP1_UBTF.p2 | 2.986 |
HMGB2
HMGB3 (HMG2A, MGC90319) SSRP1 (FACT80) UBTF (UBF, NOR-90) HBP1 |

|
|
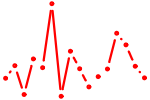
| MYB.p2 | 2.985 |
MYB
(c-myb)
|
|
|
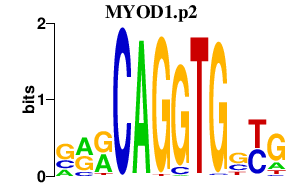
| MYOD1.p2 | 2.950 |
MYOD1
(PUM, MYOD, bHLHc1)
|
|
|
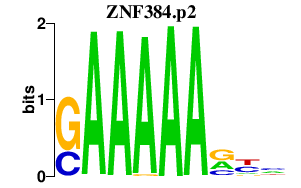
| ZNF384.p2 | 2.883 |
ZNF384
(CAGH1A, CIZ, NMP4, NP)
|
|
|
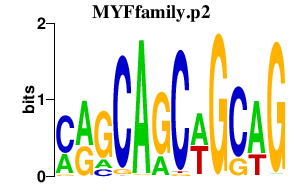
| MYFfamily.p2 | 2.828 |
MYOD1
(PUM, MYOD, bHLHc1)
MYF6 (MRF4, bHLHc4) MYF5 (bHLHc2) MYOG (bHLHc3) |
|
|
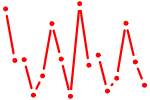
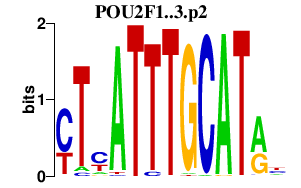
| POU2F1..3.p2 | 2.819 |
POU2F2
(OCT2)
POU2F1 (OCT1) POU2F3 (OCT11, PLA-1, Skn-1a, Epoc-1) |
|
|
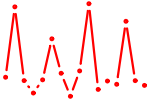
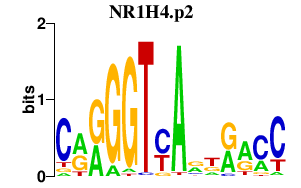
| NR1H4.p2 | 2.783 |
NR1H4
(FXR, RIP14, HRR1, HRR-1)
|
|
|
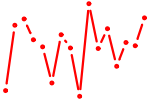
| TLX1..3_NFIC{dimer}.p2 | 2.742 |
TLX1
TLX3 (RNX) |
|
|
| POU5F1.p2 | 2.720 |
POU5F1
(OCT3, Oct4, MGC22487)
|
|

|
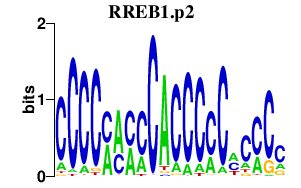
| RREB1.p2 | 2.719 |
RREB1
(HNT)
|
|
|
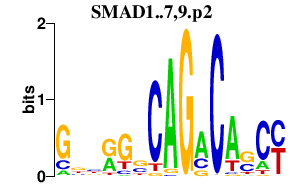
| SMAD1..7,9.p2 | 2.705 |
SMAD5
(Dwfc, JV5-1)
SMAD1 (MADR1, JV4-1) SMAD3 (JV15-2, HsT17436) SMAD4 (DPC4) SMAD2 (MADR2, JV18-1) SMAD6 (HsT17432) SMAD7 SMAD9 |
|
|
| NR5A1,2.p2 | 2.680 |
NR5A1
(FTZ1, SF-1, ELP, AD4BP)
NR5A2 (FTZ-F1beta, hB1F, LRH-1, FTZ-F1, hB1F-2, B1F2) |
|
|
| PRDM1.p3 | 2.645 |
PRDM1
(PRDI-BF1)
|

|
|
| GTF2I.p2 | 2.639 |
GTF2I
(TFII-I, BAP-135, SPIN, BTKAP1, DIWS, IB291)
|

|
|
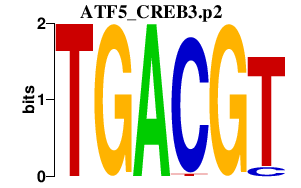
| ATF5_CREB3.p2 | 2.579 |
ATF5
CREB3 (LZIP, Luman) |
|
|
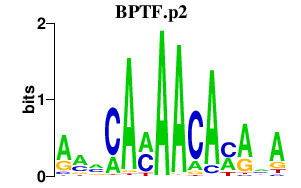
| BPTF.p2 | 2.567 |
BPTF
(FAC1, NURF301)
|
|
|
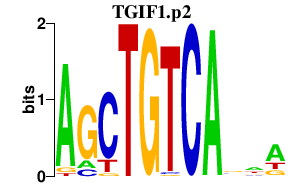
| TGIF1.p2 | 2.543 |
TGIF1
|
|
|
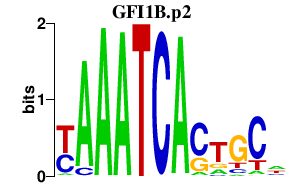
| GFI1B.p2 | 2.539 |
GFI1B
|

|
|
| NFATC1..3.p2 | 2.530 |
NFATC3
(NFAT4, NFATX)
NFATC2 (NF-ATP, NFATp, NFAT1) NFATC1 (NF-ATC, NFATc, NFAT2) |
|

|
| NFE2L2.p2 | 2.512 |
NFE2L2
(NRF2)
|
|
|
| ZIC1..3.p2 | 2.511 |
ZIC1
(ZIC, ZNF201)
ZIC2 (HPE5) ZIC3 (HTX, ZNF203) |

|
|
| PAX5.p2 | 2.501 |
PAX5
(BSAP)
|
|
|
| EP300.p2 | 2.500 |
EP300
(p300, KAT3B)
|
|
|
| ONECUT1,2.p2 | 2.488 |
ONECUT2
(OC-2)
ONECUT1 (HNF-6) |

|
|
| PATZ1.p2 | 2.439 |
PATZ1
(MAZR, dJ400N23, ZBTB19, ZSG, RIAZ, PATZ)
|
|
|
| ESRRA.p2 | 2.426 |
ESRRA
(ERR1, ERRalpha, NR3B1, ERRa)
|
|

|
| ZFP161.p2 | 2.419 |
ZFP161
(ZNF478, ZBTB14)
|
|
|
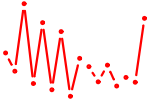
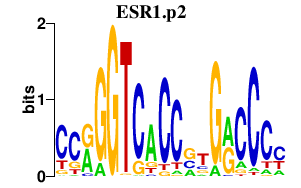
| ESR1.p2 | 2.418 |
ESR1
(NR3A1, Era)
|
|
|
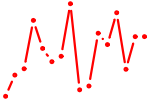
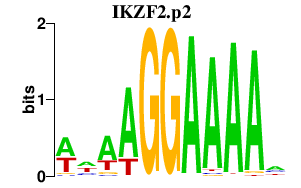
| IKZF2.p2 | 2.380 |
IKZF2
(Helios)
|
|
|
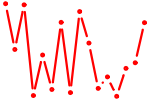
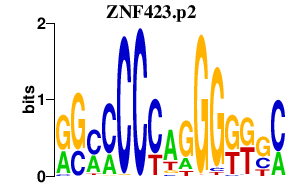
| ZNF423.p2 | 2.345 |
ZNF423
(KIAA0760, OAZ, Roaz, Ebfaz, Zfp104)
|
|
|
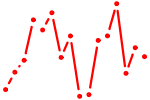
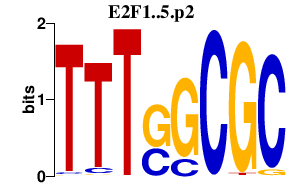
| E2F1..5.p2 | 2.344 |
E2F4
(E2F-4)
E2F5 E2F2 (E2F-2) E2F1 (RBP3) E2F3 |
|
|
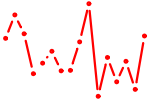
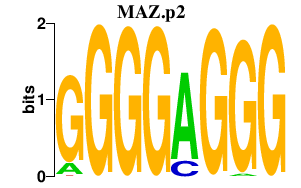
| MAZ.p2 | 2.333 |
MAZ
(ZF87, Pur-1, Zif87, ZNF801)
|
|
|
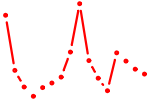
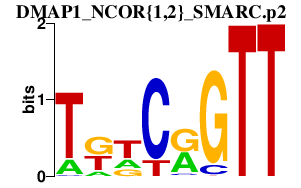
| DMAP1_NCOR{1,2}_SMARC.p2 | 2.316 |
SMARCA1
(SNF2LB, NURF140, ISWI, SWI)
SMARCC2 (BAF170, Rsc8, CRACC2) SMARCA5 (hSNF2H, hISWI, ISWI) NCOR1 (N-CoR, hCIT529I10, TRAC1, hN-CoR, KIAA1047, MGC104216) NCOR2 (SMRT, SMRTE, TRAC-1, CTG26, TNRC14) DMAP1 (DNMAP1, FLJ11543, KIAA1425, DNMTAP1, EAF2, SWC4) |
|
|
| TFDP1.p2 | 2.309 |
TFDP1
(Dp-1, DRTF1, DP1)
|
|

|
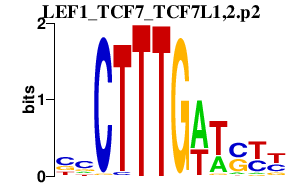
| LEF1_TCF7_TCF7L1,2.p2 | 2.224 |
TCF7
(TCF-1)
TCF7L2 (TCF-4) LEF1 (TCF1ALPHA) TCF7L1 |
|
|
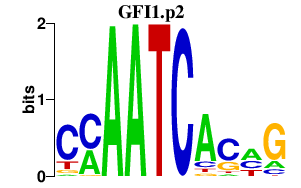
| GFI1.p2 | 2.166 |
GFI1
|

|
|
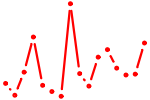
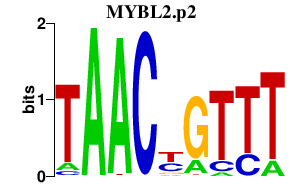
| MYBL2.p2 | 2.151 |
MYBL2
(BMYB, B-MYB)
|
|
|
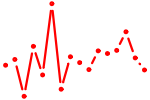
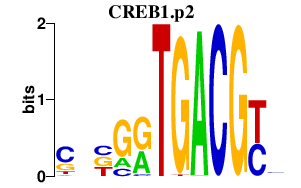
| CREB1.p2 | 2.151 |
CREB1
|
|
|
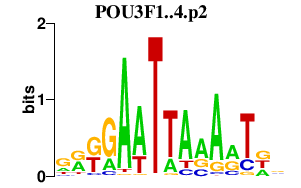
| POU3F1..4.p2 | 2.148 |
POU3F1
(OCT6, SCIP)
POU3F3 (BRN1, OTF8) POU3F2 (POUF3, BRN2, OCT7) POU3F4 (BRN4, OTF9) |
|
|
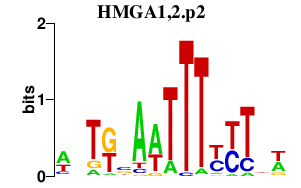
| HMGA1,2.p2 | 2.147 |
HMGA1
HMGA2 (BABL, LIPO) |
|
|
| SP1.p2 | 2.143 |
SP1
|

|
|
| FOX{I1,J2}.p2 | 2.117 |
FOXI1
(FREAC6)
FOXJ2 (FHX) |

|
|
| ARNT_ARNT2_BHLHB2_MAX_MYC_USF1.p2 | 2.117 |
ARNT
(HIF-1beta, bHLHe2)
MAX (bHLHd4) MYC (c-Myc, bHLHe39) USF1 (UEF, MLTFI, bHLHb11) BHLHB2 (DEC1, bHLHe40) ARNT2 (KIAA0307, bHLHe1) |
|
|
| EGR1..3.p2 | 2.105 |
EGR3
(PILOT)
EGR1 (TIS8, G0S30, NGFI-A, KROX-24, ZIF-268, AT225, ZNF225) EGR2 |
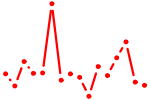
|
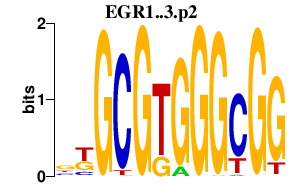
|
| HLF.p2 | 2.098 |
HLF
(MGC33822)
|

|
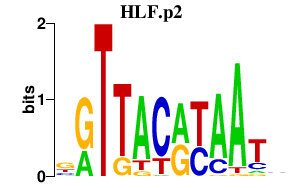
|
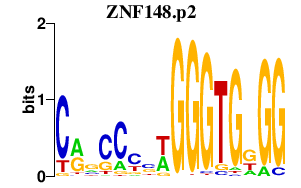
| ZNF148.p2 | 2.095 |
ZNF148
(BERF-1, ZBP-89, BFCOL1, HT-BETA, ZFP148, pHZ-52)
|
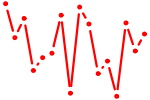
|
|
| AHR_ARNT_ARNT2.p2 | 2.088 |
ARNT
(HIF-1beta, bHLHe2)
AHR ARNT2 (KIAA0307, bHLHe1) |
|

|
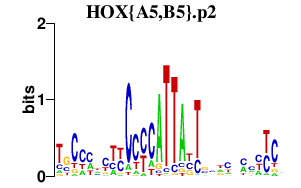
| HOX{A5,B5}.p2 | 2.087 |
HOXB5
HOXA5 |
|
|
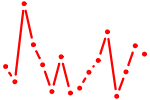
| REST.p3 | 2.084 |
REST
(NRSF, XBR)
|
|
|
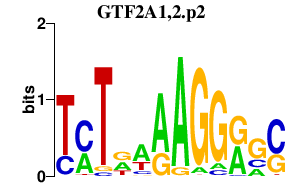
| GTF2A1,2.p2 | 2.074 |
GTF2A1
(TFIIA)
GTF2A2 (TFIIA, HsT18745) |

|
|
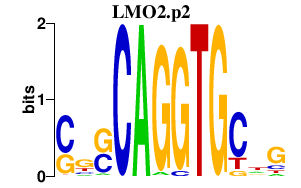
| LMO2.p2 | 2.059 |
LMO2
(TTG2, RHOM2, RBTN2)
|
|
|
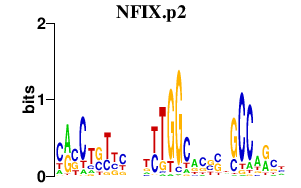
| NFIX.p2 | 2.041 |
NFIX
(NF1A)
|
|
|
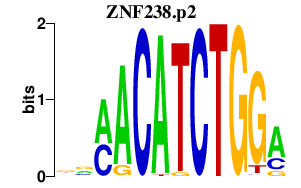
| ZNF238.p2 | 2.027 |
ZNF238
(C2H2-171, TAZ-1, RP58, ZBTB18)
|
|
|
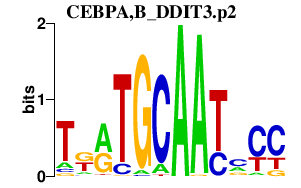
| CEBPA,B_DDIT3.p2 | 2.027 |
DDIT3
(CHOP10, GADD153, CHOP)
CEBPA (C/EBP-alpha) CEBPB (LAP, CRP2, NFIL6, IL6DBP) |
|
|
| GATA1..3.p2 | 2.007 |
GATA1
(ERYF1, NFE1)
GATA2 (NFE1B) GATA3 (HDR) |
|

|
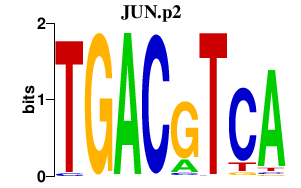
| JUN.p2 | 2.000 |
JUN
(c-Jun, AP-1)
JUND (AP-1) JUNB |
|
|
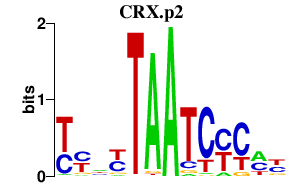
| CRX.p2 | 1.959 |
CRX
(CRD, LCA7, OTX3)
|
|
|
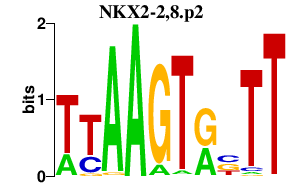
| NKX2-2,8.p2 | 1.952 |
NKX2-2
(NKX2.2)
NKX2-8 (NKX2.8, Nkx2-9) |
|
|
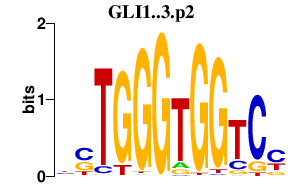
| GLI1..3.p2 | 1.947 |
GLI1
GLI2 (THP2, HPE9, THP1) GLI3 (PAP-A, PAPA, PAPA1, PAPB, ACLS, PPDIV) |
|
|
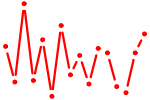
| HMX1.p2 | 1.935 |
HMX1
(H6, NKX5-3)
|
|
|
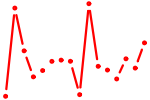
| MSX1,2.p2 | 1.926 |
MSX1
(HYD1, OFC5)
MSX2 (CRS2, FPP, HOX8, MSH, PFM) |
|

|
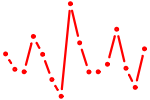
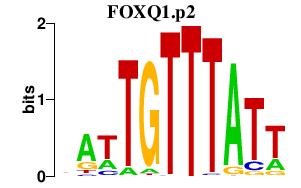
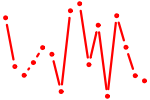
| FOXQ1.p2 | 1.912 |
FOXQ1
(HFH1)
|
|
|
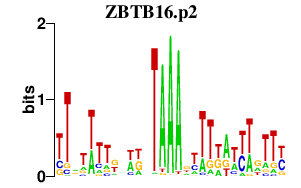
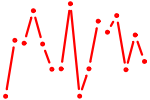
| ZBTB16.p2 | 1.900 |
ZBTB16
(PLZF)
|
|
|
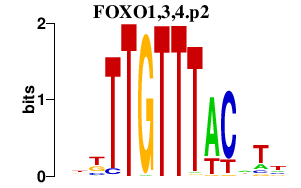
| FOXO1,3,4.p2 | 1.880 |
FOXO1
(FKH1)
FOXO4 (AFX1) FOXO3 (AF6q21, FOXO2) |
|
|
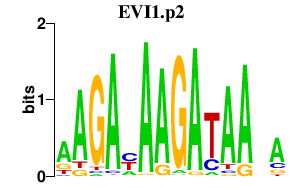
| EVI1.p2 | 1.848 |
EVI1
(MDS1-EVI1, PRDM3)
|
|
|
| TFAP4.p2 | 1.844 |
TFAP4
(AP-4)
|
|
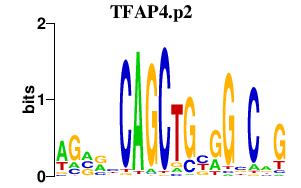
|
| SRF.p3 | 1.817 |
SRF
(MCM1)
|
|
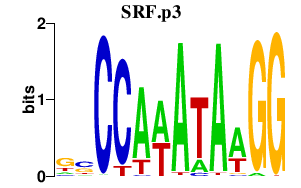
|
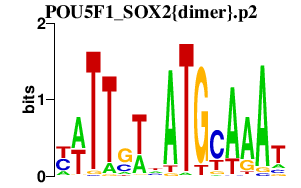
| POU5F1_SOX2{dimer}.p2 | 1.816 |
POU5F1
(OCT3, Oct4, MGC22487)
SOX2 |
|
|
| NFE2L1.p2 | 1.799 |
NFE2L1
(NRF1, LCR-F1, FLJ00380, TCF11)
|
|

|
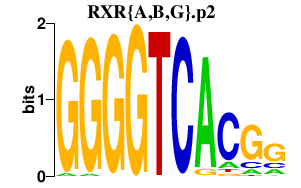
| RXR{A,B,G}.p2 | 1.790 |
RXRA
(NR2B1)
RXRB (NR2B2, H-2RIIBP, RCoR-1) RXRG (NR2B3) |
|
|
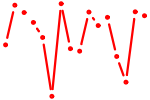
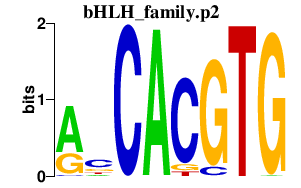
| bHLH_family.p2 | 1.786 |
ARNTL
(MOP3, JAP3, BMAL1, PASD3, bHLHe5)
MITF (WS2A, MI, bHLHe32) MXI1 (MXD2, MAD2, MXI, bHLHc11) TFE3 (TFEA, bHLHe33) CLOCK (KIAA0334, KAT13D, bHLHe8) OLIG2 (RACK17, OLIGO2, bHLHe19) MXD4 (MAD4, MSTP149, MST149, bHLHc12) HEY2 (bHLHb32) HEY1 (HESR-1, CHF2, HESR1, HRT-1, CHF-2, HERP2, BHLHb31) HEYL (bHLHb33) MLXIPL (WS-bHLH, MIO, CHREBP, MONDOB) HES6 (bHLHc23) MXD3 (MAD3, bHLHc13) ARNTL2 (BMAL2, MOP9, CLIF, PASD9, bHLHe6) ID1 (dJ857M17.1.2, bHLHb24) MNT (ROX, MXD6, MAD6, bHLHd3) OLIG1 (BHLHB6, bHLHe21) NPAS2 (MOP4, PASD4, bHLHe9) |
|
|
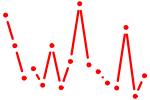
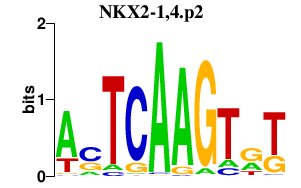
| NKX2-1,4.p2 | 1.781 |
NKX2-1
(TTF-1)
NKX2-4 (NKX2.4) |
|
|
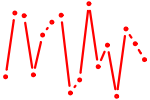
| TBP.p2 | 1.780 |
TBP
(TFIID)
|
|
|
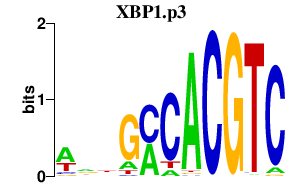
| XBP1.p3 | 1.716 |
XBP1
|
|
|
| NKX3-1.p2 | 1.683 |
NKX3-1
(NKX3.1, BAPX2)
|
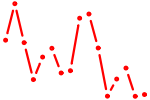
|
|
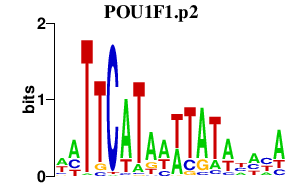
| POU1F1.p2 | 1.664 |
POU1F1
(GHF-1)
|
|
|
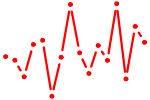
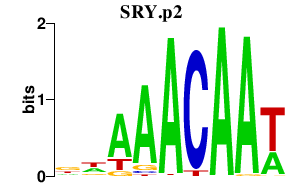
| SRY.p2 | 1.654 |
SRY
(TDF)
|
|
|
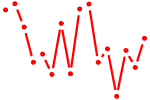
| RXRA_VDR{dimer}.p2 | 1.588 |
VDR
(NR1I1)
|
|

|
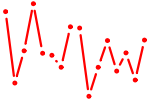
| FOXL1.p2 | 1.569 |
FOXL1
(FREAC7, FKH6)
|
|
|
| SOX{8,9,10}.p2 | 1.558 |
SOX9
(SRA1)
SOX8 SOX10 (DOM, WS4, WS2E) |
|
|
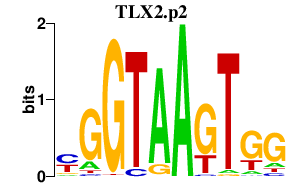
| TLX2.p2 | 1.524 |
TLX2
(Enx, Tlx2, NCX)
|
|
|
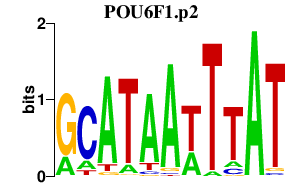
| POU6F1.p2 | 1.514 |
POU6F1
(BRN5, MPOU, TCFB1)
|
|
|
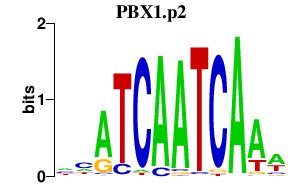
| PBX1.p2 | 1.506 |
PBX1
|
|
|
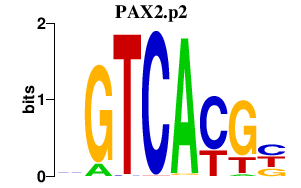
| PAX2.p2 | 1.505 |
PAX2
|
|
|
| ZBTB6.p2 | 1.502 |
ZBTB6
(ZID)
|
|
|
| NKX3-2.p2 | 1.492 |
NKX3-2
(NKX3B, NKX3.2)
|
|
|
| FOX{F1,F2,J1}.p2 | 1.458 |
FOXF2
(FREAC2)
FOXJ1 (HFH-4, HFH4) FOXF1 (FREAC1) |

|
|
| TAL1_TCF{3,4,12}.p2 | 1.457 |
TCF3
(E2A, ITF1, MGC129647, MGC129648, bHLHb21)
TCF4 (SEF2-1B, ITF2, bHLHb19) TAL1 (SCL, bHLHa17) TCF12 (HEB, HTF4, HsT17266, bHLHb20) |
|

|
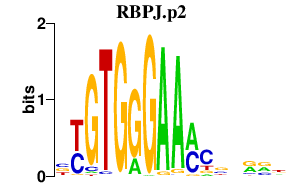
| RBPJ.p2 | 1.452 |
RBPJ
(SUH, IGKJRB, RBPJK, KBF2, RBP-J, CBF1)
|

|
|
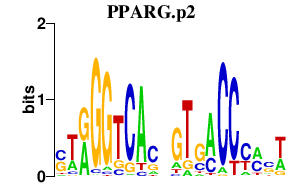
| PPARG.p2 | 1.427 |
PPARG
(PPARG1, PPARG2, NR1C3, PPARgamma)
|
|
|
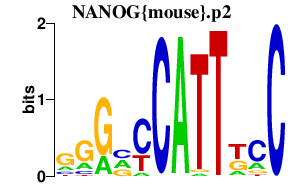
| NANOG{mouse}.p2 | 1.419 |
NANOG
(FLJ12581, FLJ40451)
|
|
|
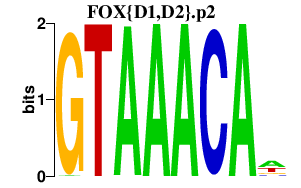
| FOX{D1,D2}.p2 | 1.394 |
FOXD2
(FREAC9)
FOXD1 (FREAC4) |
|
|
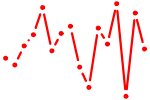
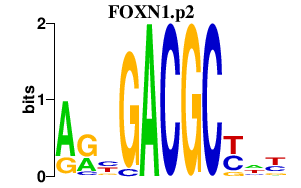
| FOXN1.p2 | 1.358 |
FOXN1
(FKHL20)
|
|
|
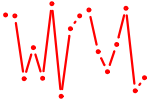
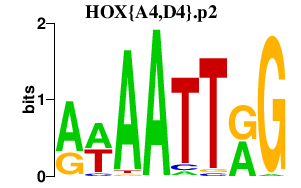
| HOX{A4,D4}.p2 | 1.348 |
HOXA4
HOXD4 |
|
|
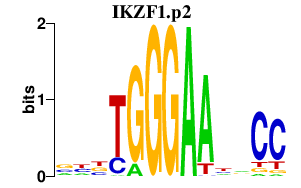
| IKZF1.p2 | 1.307 |
IKZF1
(hIk-1, LyF-1, Hs.54452, IKAROS)
|
|
|
| SOX5.p2 | 1.299 |
SOX5
(L-SOX5, MGC35153)
|
|

|
| HSF1,2.p2 | 1.294 |
HSF1
(HSTF1)
HSF2 |
|
|
| FOXP3.p2 | 1.288 |
FOXP3
(JM2, XPID, AIID, PIDX, DIETER, SCURFIN)
|
|
|
| NKX6-1,2.p2 | 1.286 |
NKX6-1
(Nkx6.1)
NKX6-2 (NKX6B, GTX, NKX6.1) |
|
|
| FOXD3.p2 | 1.281 |
FOXD3
(Genesis, HFH2)
|
|

|
| BACH2.p2 | 1.280 |
BACH2
|
|
|
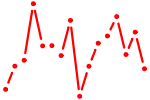
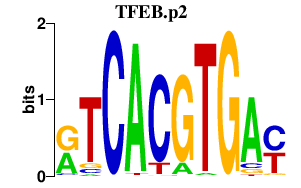
| TFEB.p2 | 1.276 |
TFEB
|
|
|
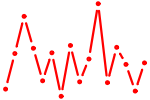
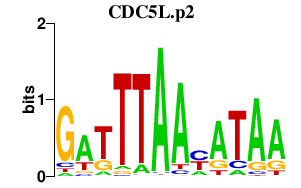
| CDC5L.p2 | 1.271 |
CDC5L
(PCDC5RP, hCDC5, CEF1)
|
|
|
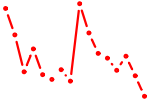
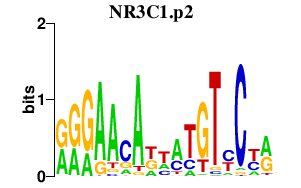
| NR3C1.p2 | 1.266 |
NR3C1
(GR)
|
|
|
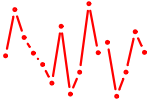
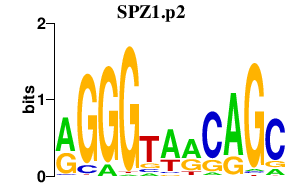
| SPZ1.p2 | 1.259 |
SPZ1
(NYD-TSP1, FLJ25709)
|
|
|
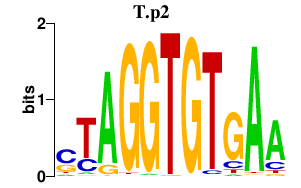
| T.p2 | 1.252 |
T
|
|
|
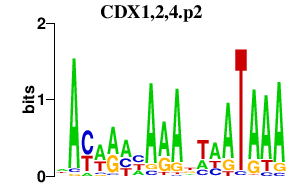
| CDX1,2,4.p2 | 1.250 |
CDX1
CDX2 CDX4 |
|
|
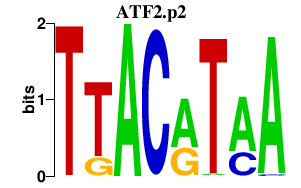
| ATF2.p2 | 1.250 |
ATF2
(TREB7, CRE-BP1, HB16)
|
|
|
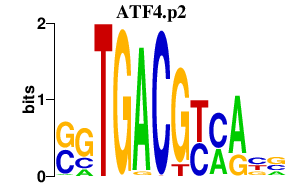
| ATF4.p2 | 1.245 |
ATF4
(TAXREB67, CREB-2)
|
|
|
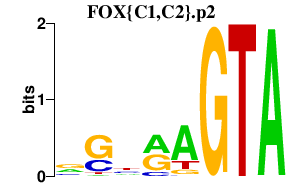
| FOX{C1,C2}.p2 | 1.238 |
FOXC1
(FREAC3, ARA, IGDA, IHG1)
FOXC2 (MFH-1) |
|
|
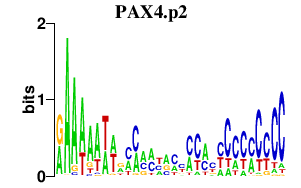
| PAX4.p2 | 1.228 |
PAX4
|
|
|
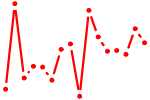
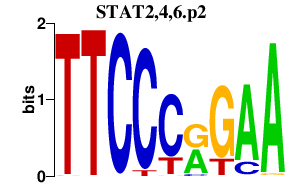
| STAT2,4,6.p2 | 1.215 |
STAT2
(STAT113)
STAT4 STAT6 (D12S1644, IL-4-STAT) |
|
|
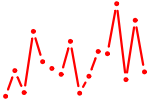
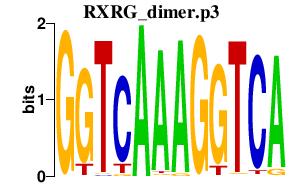
| RXRG_dimer.p3 | 1.214 |
NR1H2
(NER, NER-I, RIP15, LXR-b)
RXRA (NR2B1) PPARG (PPARG1, PPARG2, NR1C3, PPARgamma) PPARA (hPPAR, NR1C1) RXRB (NR2B2, H-2RIIBP, RCoR-1) RXRG (NR2B3) |
|
|
| NANOG.p2 | 1.193 |
NANOG
(FLJ12581, FLJ40451)
|
|

|
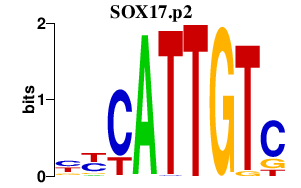
| SOX17.p2 | 1.176 |
SOX17
|
|
|
| FOSL2.p2 | 1.164 |
FOSL2
(FRA2, FLJ23306)
|
|
|
| HOXA9_MEIS1.p2 | 1.162 |
MEIS1
HOXA9 |
|
|
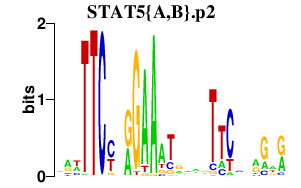
| STAT5{A,B}.p2 | 1.152 |
STAT5B
STAT5A (MGF) |
|
|
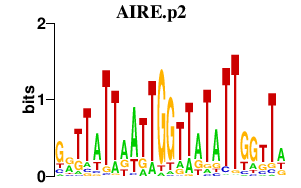
| AIRE.p2 | 1.146 |
AIRE
(PGA1, APS1)
|
|
|
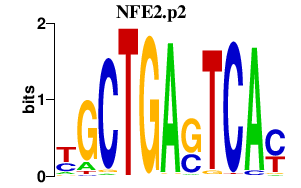
| NFE2.p2 | 1.145 |
NFE2
(NF-E2)
|
|
|
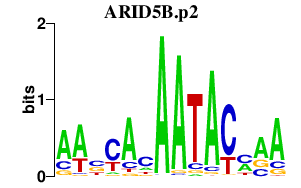
| ARID5B.p2 | 1.141 |
ARID5B
(FLJ21150, MRF2)
|
|
|
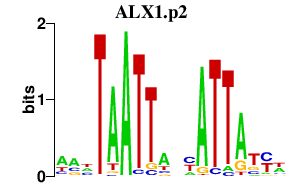
| ALX1.p2 | 1.113 |
ALX1
|
|
|
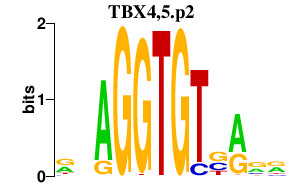
| TBX4,5.p2 | 1.107 |
TBX5
TBX4 |
|
|
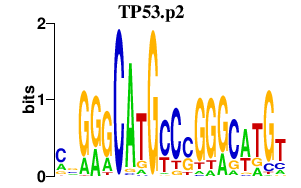
| TP53.p2 | 1.103 |
TP53
(p53, LFS1)
|
|
|
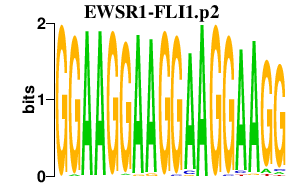
| EWSR1-FLI1.p2 | 1.100 |
FLI1
(EWSR2, SIC-1)
EWSR1 (EWS, K984G1.4) |
|
|
| NKX2-3_NKX2-5.p2 | 1.078 |
NKX2-5
(CSX1, NKX2.5, NKX4-1)
NKX2-3 (NKX2.3, CSX3, NKX4-3) |
|
|
| HIF1A.p2 | 1.047 |
HIF1A
(MOP1, HIF-1alpha, PASD8, HIF1)
|
|
|
| EOMES.p2 | 1.007 |
EOMES
(TBR2)
|
|
|
| CUX2.p2 | 1.002 |
CUX2
(KIAA0293, CDP2)
|
|
|
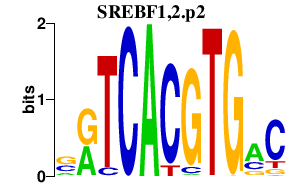
| SREBF1,2.p2 | 0.980 |
SREBF1
(SREBP1, bHLHd1)
SREBF2 (SREBP2, bHLHd2) |
|
|
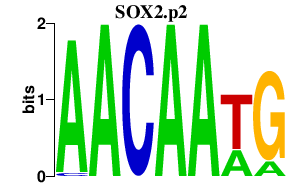
| SOX2.p2 | 0.972 |
SOX2
|
|
|
| ADNP_IRX_SIX_ZHX.p2 | 0.967 |
SIX5
ZHX1 IRX4 ADNP (KIAA0784, ADNP1) ZHX3 (KIAA0395) ZHX2 (KIAA0854) IRX5 (IRX-2a) SIX2 |
|
|
| NR6A1.p2 | 0.954 |
NR6A1
(GCNF1, RTR)
|
|
|
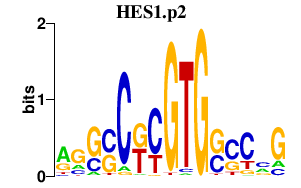
| HES1.p2 | 0.934 |
HES1
(FLJ20408, HES-1, Hes1, bHLHb39)
|
|
|
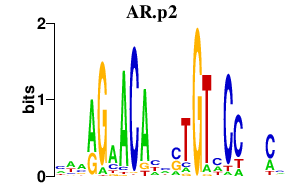
| AR.p2 | 0.926 |
AR
(AIS, NR3C4, SMAX1, HUMARA)
|
|
|
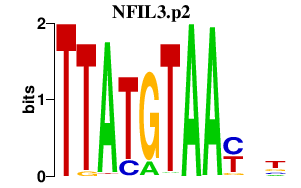
| NFIL3.p2 | 0.889 |
NFIL3
(E4BP4, NFIL3A, NF-IL3A)
|
|
|
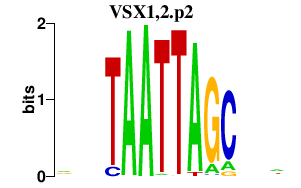
| VSX1,2.p2 | 0.854 |
VSX2
(RET1)
VSX1 (PPD) |
|
|
| LHX3,4.p2 | 0.839 |
LHX3
LHX4 (Gsh4) |
|
|
| ATF6.p2 | 0.834 |
ATF6
(ATF6A)
|
|

|
| STAT1,3.p3 | 0.737 |
STAT1
(STAT91, ISGF-3)
STAT3 (APRF) |
|
|
| PAX6.p2 | 0.704 |
PAX6
(D11S812E, AN, WAGR)
|
|

|
| PDX1.p2 | 0.651 |
PDX1
(IDX-1, STF-1, PDX-1, MODY4)
|
|
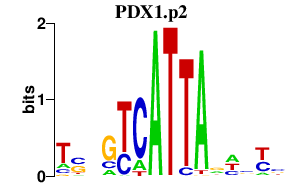
|
| RORA.p2 | 0.525 |
RORA
(RZRA, ROR1, ROR2, ROR3, NR1F1)
|
|
|
| GZF1.p2 | 0.459 |
GZF1
(dJ322G13.2, ZBTB23)
|

|
|
| PAX8.p2 | 0.281 |
PAX8
|
|
|
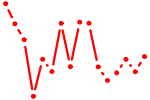
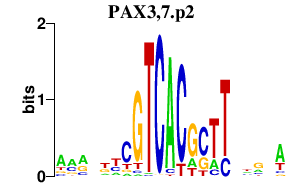
| PAX3,7.p2 | 0.227 |
PAX7
(Hup1)
PAX3 (HUP2) |
|
|
All samples sorted alphabetically
| Your email address: | |
| Project name: |
| Sample name | Condition | Replicate/Batch |
|---|---|---|
| Rep1_GM12878 | ||
| Rep1_HepG2 | ||
| Rep1_HMEC | ||
| Rep1_HSMM | ||
| Rep1_Huvec | ||
| Rep1_K562 | ||
| Rep1_NHEK | ||
| Rep1_NHLF | ||
| Rep2_GM12878 | ||
| Rep2_HepG2 | ||
| Rep2_HMEC | ||
| Rep2_HSMM | ||
| Rep2_Huvec | ||
| Rep2_K562 | ||
| Rep2_NHEK | ||
| Rep2_NHLF |