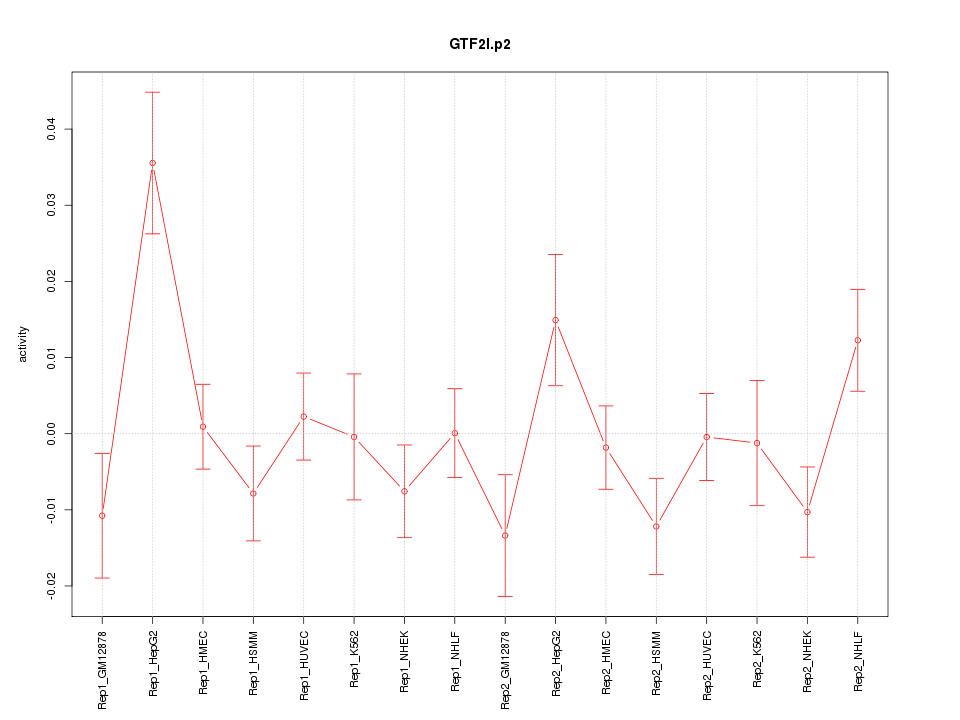
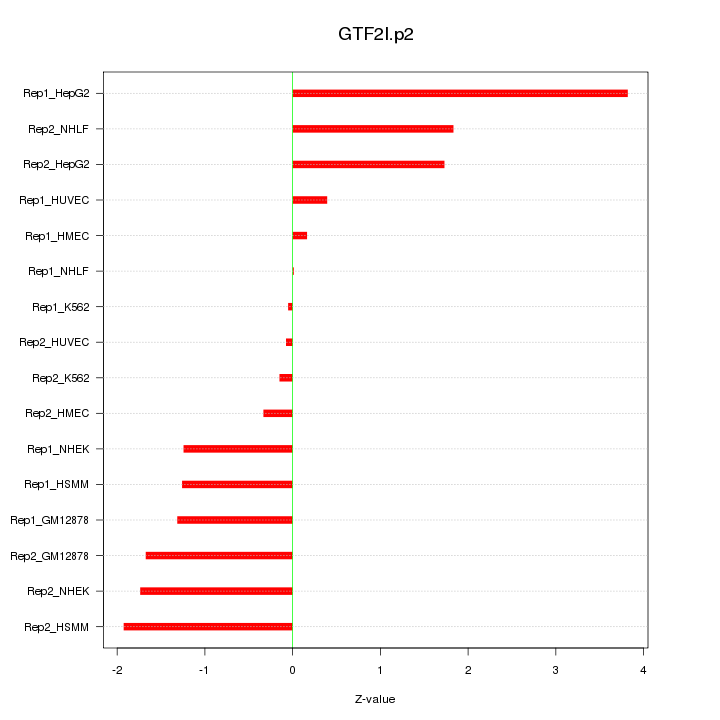
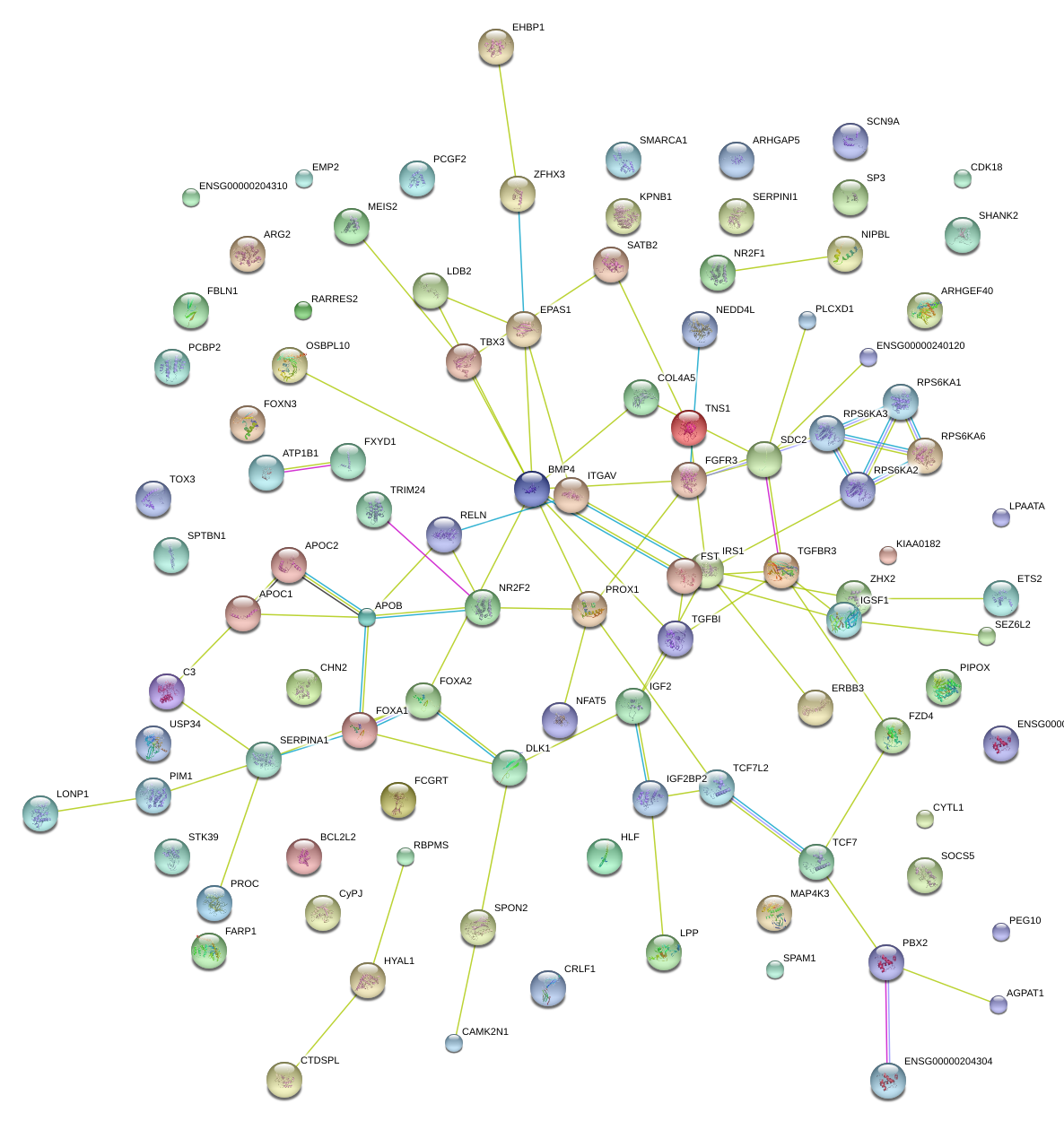
|
chrY_+_148060
|
5.210
|
NM_018390
|
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1
|
|
chrX_+_198060
|
5.145
|
NM_018390
|
PLCXD1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1
|
|
chr20_-_22564893
|
4.566
|
|
FOXA2
|
forkhead box A2
|
|
chr19_+_45452427
|
3.756
|
|
APOC2
|
apolipoprotein C-II
|
|
chr11_-_2159472
|
2.735
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr1_-_20812727
|
2.504
|
NM_018584
|
CAMK2N1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr20_-_22565050
|
2.363
|
NM_021784
|
FOXA2
|
forkhead box A2
|
|
chr12_-_115121939
|
2.315
|
NM_005996
NM_016569
|
TBX3
|
T-box 3
|
|
chr14_-_54423448
|
2.228
|
NM_001202
NM_130850
|
BMP4
|
bone morphogenetic protein 4
|
|
chr10_+_114709963
|
2.074
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr11_-_2160199
|
2.072
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr8_+_97506141
|
2.050
|
|
SDC2
|
syndecan 2
|
|
chr11_-_2159925
|
2.049
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr1_-_92351476
|
2.036
|
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr2_-_39664430
|
2.032
|
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr16_-_29910340
|
1.961
|
NM_001114099
NM_001114100
NM_001243332
NM_001243333
NM_012410
NM_201575
|
SEZ6L2
|
seizure related 6 homolog (mouse)-like 2
|
|
chr2_+_128175989
|
1.904
|
NM_000312
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chr2_+_128176019
|
1.898
|
|
PROC
|
protein C (inactivator of coagulation factors Va and VIIIa)
|
|
chr5_+_92918924
|
1.844
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr5_+_92920592
|
1.747
|
|
NR2F1
NR2F2
|
nuclear receptor subfamily 2, group F, member 1
nuclear receptor subfamily 2, group F, member 2
|
|
chr19_+_50016435
|
1.652
|
NM_004107
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr11_-_2159784
|
1.651
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr14_+_32546445
|
1.590
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr2_+_62932786
|
1.581
|
NM_001142616
NM_015252
|
EHBP1
|
EH domain binding protein 1
|
|
chr2_-_39664291
|
1.571
|
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr2_-_227663454
|
1.544
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr8_+_97506071
|
1.534
|
|
SDC2
|
syndecan 2
|
|
chr19_-_6720585
|
1.529
|
NM_000064
|
C3
|
complement component 3
|
|
chr16_-_73092533
|
1.521
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr8_+_97506020
|
1.503
|
|
SDC2
|
syndecan 2
|
|
chr17_-_36904545
|
1.485
|
NM_007144
|
PCGF2
|
polycomb group ring finger 2
|
|
chr2_-_218843541
|
1.476
|
|
TNS1
|
tensin 1
|
|
chr1_-_92351613
|
1.470
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr16_+_69599869
|
1.456
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr16_+_69599868
|
1.440
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr8_+_97505895
|
1.430
|
|
SDC2
|
syndecan 2
|
|
chr7_+_29234111
|
1.371
|
NM_004067
|
CHN2
|
chimerin (chimaerin) 2
|
|
chrX_-_128657453
|
1.355
|
NM_003069
NM_139035
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr7_-_150038689
|
1.351
|
NM_002889
|
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2
|
|
chr3_-_50349811
|
1.345
|
NM_153281
|
HYAL1
|
hyaluronoglucosaminidase 1
|
|
chr14_+_21538339
|
1.333
|
NM_018071
|
ARHGEF40
|
Rho guanine nucleotide exchange factor (GEF) 40
|
|
chrX_-_128657440
|
1.313
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chrX_-_130423387
|
1.304
|
NM_001170961
NM_001170962
NM_001555
NM_205833
|
IGSF1
|
immunoglobulin superfamily, member 1
|
|
chrX_-_20284910
|
1.299
|
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr4_-_1166959
|
1.286
|
NM_001128325
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr6_+_37137882
|
1.266
|
NM_001243186
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr3_+_187871473
|
1.237
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr19_+_35629727
|
1.220
|
NM_021902
|
FXYD1
|
FXYD domain containing ion transport regulator 1
|
|
chrX_-_130423199
|
1.188
|
|
IGSF1
|
immunoglobulin superfamily, member 1
|
|
chr22_-_36236330
|
1.173
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chrX_-_128657381
|
1.169
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr14_-_54421269
|
1.137
|
NM_130851
|
BMP4
|
bone morphogenetic protein 4
|
|
chr5_+_135364583
|
1.128
|
NM_000358
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr11_-_86666211
|
1.120
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chrX_-_20285141
|
1.119
|
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr19_+_45417920
|
1.081
|
NM_001645
|
APOC1
|
apolipoprotein C-I
|
|
chr8_+_97505876
|
1.078
|
NM_002998
|
SDC2
|
syndecan 2
|
|
chr3_-_185542807
|
1.076
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr21_+_40177754
|
1.075
|
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr2_-_169103886
|
1.062
|
NM_013233
|
STK39
|
serine threonine kinase 39
|
|
chr2_-_21266809
|
1.061
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr13_+_98795710
|
1.061
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr2_+_54683276
|
1.052
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr15_-_37392322
|
1.031
|
|
MEIS2
|
Meis homeobox 2
|
|
chr16_-_30934437
|
1.029
|
|
FBXL19-AS1
|
FBXL19 antisense RNA 1 (non-protein coding)
|
|
chr14_+_101193190
|
1.029
|
NM_003836
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr18_+_55711890
|
1.028
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr2_-_61697899
|
1.024
|
|
USP34
|
ubiquitin specific peptidase 34
|
|
chr5_+_36876838
|
1.014
|
NM_015384
NM_133433
|
NIPBL
|
Nipped-B homolog (Drosophila)
|
|
chr14_+_101193275
|
1.014
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr1_+_214161279
|
1.009
|
|
PROX1
|
prospero homeobox 1
|
|
chr2_-_39664129
|
1.007
|
NM_003618
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr17_+_45727303
|
1.000
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr17_+_45727558
|
0.987
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr2_+_46926049
|
0.984
|
NM_014011
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr16_+_69600110
|
0.980
|
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr14_+_23775970
|
0.973
|
NM_001199864
NM_004050
NM_001199839
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 readthrough
BCL2-like 2
|
|
chr17_-_7155258
|
0.972
|
NM_015343
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1
|
|
chr2_-_200322699
|
0.952
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr16_+_85645014
|
0.951
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr3_+_37903643
|
0.947
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr15_-_37392358
|
0.945
|
NM_170674
NM_170675
NM_170676
NM_170677
|
MEIS2
|
Meis homeobox 2
|
|
chr14_+_68086633
|
0.944
|
|
ARG2
|
arginase, type II
|
|
chr19_-_18717577
|
0.941
|
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr6_-_32143886
|
0.939
|
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr17_+_53342356
|
0.939
|
|
HLF
|
hepatic leukemia factor
|
|
chr2_-_167232464
|
0.938
|
NM_002977
|
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit
|
|
chr3_+_167453431
|
0.933
|
NM_001122752
NM_005025
|
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1
|
|
chr8_+_123793988
|
0.927
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr5_+_36876899
|
0.922
|
|
NIPBL
|
Nipped-B homolog (Drosophila)
|
|
chr22_-_36236264
|
0.908
|
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr2_-_174828775
|
0.908
|
NM_001017371
|
SP3
|
Sp3 transcription factor
|
|
chr14_-_89883306
|
0.902
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr2_+_46926323
|
0.900
|
NM_144949
|
SOCS5
|
suppressor of cytokine signaling 5
|
|
chr21_+_40177847
|
0.899
|
NM_005239
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr2_-_21266893
|
0.898
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr14_+_101193313
|
0.896
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr4_-_16900348
|
0.885
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr4_-_1166547
|
0.881
|
NM_012445
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr6_-_32143795
|
0.875
|
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr2_-_21266915
|
0.870
|
NM_000384
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr22_-_36236421
|
0.865
|
NM_001031695
NM_001082576
NM_001082577
NM_014309
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr16_-_10674443
|
0.855
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr14_-_38064440
|
0.844
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr2_+_187454724
|
0.842
|
NM_001145000
NM_002210
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chrX_+_49644469
|
0.841
|
NM_001145073
|
USP27X
|
ubiquitin specific peptidase 27, X-linked
|
|
chr8_+_30241943
|
0.824
|
NM_001008710
NM_001008711
NM_001008712
NM_006867
|
RBPMS
|
RNA binding protein with multiple splicing
|
|
chr12_+_53845884
|
0.824
|
NM_001098620
NM_001128911
NM_001128912
NM_001128913
NM_001128914
NM_005016
NM_031989
|
PCBP2
|
poly(rC) binding protein 2
|
|
chr14_-_94856926
|
0.821
|
NM_001002235
NM_001002236
NM_001127700
NM_001127701
NM_001127702
NM_001127703
NM_001127704
NM_001127705
NM_001127706
NM_001127707
|
SERPINA1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1
|
|
chr2_-_227664330
|
0.813
|
|
IRS1
|
insulin receptor substrate 1
|
|
chr17_+_53342320
|
0.808
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr19_+_50016490
|
0.804
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr7_+_94285620
|
0.802
|
NM_001040152
NM_001184961
NM_001184962
NM_015068
|
PEG10
|
paternally expressed 10
|
|
chr1_+_214161859
|
0.796
|
NM_002763
|
PROX1
|
prospero homeobox 1
|
|
chr3_-_32022734
|
0.796
|
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr4_+_1795004
|
0.795
|
NM_000142
NM_001163213
NM_022965
|
FGFR3
|
fibroblast growth factor receptor 3
|
|
chr16_-_52580805
|
0.791
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr7_-_103629962
|
0.787
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr3_-_32023237
|
0.784
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr1_+_169075869
|
0.781
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr22_+_45898690
|
0.778
|
NM_001996
NM_006485
NM_006486
NM_006487
|
FBLN1
|
fibulin 1
|
|
chr1_+_205473768
|
0.773
|
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr5_+_52776408
|
0.769
|
|
FST
|
follistatin
|
|
chr17_+_27370155
|
0.768
|
|
PIPOX
|
pipecolic acid oxidase
|
|
chr11_-_70672644
|
0.768
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr6_-_32157511
|
0.768
|
NM_002586
|
PBX2
|
pre-B-cell leukemia homeobox 2
|
|
chr12_+_56473944
|
0.762
|
|
ERBB3
|
v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian)
|
|
chr10_+_114710785
|
0.757
|
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr7_+_138144847
|
0.752
|
NM_003852
NM_015905
|
TRIM24
|
tripartite motif containing 24
|
|
chrX_+_107683015
|
0.748
|
NM_000495
NM_033380
|
COL4A5
|
collagen, type IV, alpha 5
|
|
chrX_+_152954965
|
0.747
|
NM_001142806
|
SLC6A8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8
|
|
chr20_+_8112762
|
0.745
|
NM_015192
NM_182734
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific)
|
|
chr19_-_18717631
|
0.743
|
NM_004750
|
CRLF1
|
cytokine receptor-like factor 1
|
|
chr2_-_136743189
|
0.740
|
NM_001349
|
DARS
|
aspartyl-tRNA synthetase
|
|
chr6_-_134497069
|
0.737
|
NM_001143678
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr19_-_48894755
|
0.732
|
NM_006801
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1
|
|
chr5_+_52776479
|
0.730
|
|
FST
|
follistatin
|
|
chr7_+_45927958
|
0.730
|
NM_000596
|
IGFBP1
|
insulin-like growth factor binding protein 1
|
|
chr17_+_27369917
|
0.729
|
NM_016518
|
PIPOX
|
pipecolic acid oxidase
|
|
chr5_+_150400132
|
0.729
|
|
GPX3
|
glutathione peroxidase 3 (plasma)
|
|
chr7_+_94138991
|
0.723
|
NM_022900
|
CASD1
|
CAS1 domain containing 1
|
|
chr7_+_73868397
|
0.720
|
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr20_-_3154169
|
0.716
|
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr3_+_9439290
|
0.711
|
NM_001080517
|
SETD5
|
SET domain containing 5
|
|
chr20_+_44035203
|
0.704
|
NM_001048223
NM_001048224
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr12_+_3068442
|
0.704
|
NM_003213
NM_201441
NM_201443
|
TEAD4
|
TEA domain family member 4
|
|
chr1_+_205473683
|
0.703
|
NM_002596
NM_212502
NM_212503
|
CDK18
|
cyclin-dependent kinase 18
|
|
chr7_+_73868111
|
0.703
|
NM_001199207
NM_005685
NM_016328
|
GTF2IRD1
|
GTF2I repeat domain containing 1
|
|
chr5_-_172756505
|
0.703
|
NM_003714
|
STC2
|
stanniocalcin 2
|
|
chr13_-_96296919
|
0.702
|
NM_014934
NM_198968
|
DZIP1
|
DAZ interacting protein 1
|
|
chr17_-_7082654
|
0.702
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr8_+_123794043
|
0.699
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr2_+_219283809
|
0.699
|
NM_007127
|
VIL1
|
villin 1
|
|
chr1_+_204042191
|
0.693
|
NM_005686
|
SOX13
|
SRY (sex determining region Y)-box 13
|
|
chr17_+_45726838
|
0.691
|
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr17_+_70117177
|
0.690
|
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr13_+_98795468
|
0.685
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr19_+_50016500
|
0.683
|
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha
|
|
chr19_+_56652529
|
0.681
|
NM_001253792
NM_018337
|
ZNF444
|
zinc finger protein 444
|
|
chr13_+_98795410
|
0.679
|
NM_001001715
NM_005766
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr11_+_73359734
|
0.679
|
NM_001130036
|
PLEKHB1
|
pleckstrin homology domain containing, family B (evectins) member 1
|
|
chr7_-_27170351
|
0.678
|
NM_002141
|
HOXA4
|
homeobox A4
|
|
chr6_-_134496833
|
0.677
|
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr17_-_41623246
|
0.677
|
NM_001986
|
ETV4
|
ets variant 4
|
|
chr6_+_31868775
|
0.673
|
NM_001178063
|
C2
|
complement component 2
|
|
chr19_-_48894615
|
0.665
|
|
KDELR1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1
|
|
chr17_-_53499055
|
0.665
|
NM_012329
|
MMD
|
monocyte to macrophage differentiation-associated
|
|
chr3_-_185542744
|
0.663
|
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr11_-_35441099
|
0.662
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr2_-_174829108
|
0.661
|
|
SP3
|
Sp3 transcription factor
|
|
chr20_+_306205
|
0.659
|
NM_006943
|
SOX12
|
SRY (sex determining region Y)-box 12
|
|
chr2_-_136743076
|
0.654
|
|
DARS
|
aspartyl-tRNA synthetase
|
|
chr12_-_81331487
|
0.652
|
NM_004664
|
LIN7A
|
lin-7 homolog A (C. elegans)
|
|
chr18_+_32621299
|
0.650
|
NM_014268
|
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2
|
|
chr8_-_134584012
|
0.647
|
NM_003033
NM_173344
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr8_+_123793894
|
0.646
|
NM_014943
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr10_+_21823093
|
0.642
|
NM_001195627
NM_001195628
NM_001195630
NM_004641
|
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10
|
|
chr6_+_1390068
|
0.639
|
NM_001452
|
FOXF2
|
forkhead box F2
|
|
chr6_+_33378772
|
0.633
|
NM_002636
NM_024165
|
PHF1
|
PHD finger protein 1
|
|
chr19_+_35521591
|
0.633
|
NM_001037
NM_199037
|
SCN1B
|
sodium channel, voltage-gated, type I, beta
|
|
chr1_-_227505796
|
0.633
|
NM_003607
NM_014826
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like)
|
|
chr17_+_45727274
|
0.630
|
NM_002265
|
KPNB1
|
karyopherin (importin) beta 1
|
|
chr11_+_57365026
|
0.629
|
NM_000062
|
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1
|
|
chr18_+_55711388
|
0.629
|
NM_001144967
NM_001243960
NM_015277
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr6_-_32143899
|
0.627
|
NM_006411
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr2_+_159313466
|
0.621
|
NM_001005476
NM_003628
|
PKP4
|
plakophilin 4
|
|
chr3_-_9439122
|
0.620
|
|
LOC440944
|
uncharacterized LOC440944
|
|
chr4_-_80994183
|
0.618
|
|
|
|
|
chr6_-_32143676
|
0.618
|
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chrX_+_54834775
|
0.618
|
NM_014599
|
MAGED2
|
melanoma antigen family D, 2
|
|
chr4_-_39528921
|
0.616
|
|
UGDH
|
UDP-glucose 6-dehydrogenase
|
|
chr11_-_35440514
|
0.613
|
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr1_+_116916488
|
0.610
|
NM_001160233
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide
|
|
chr20_+_34742656
|
0.608
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr17_+_40834571
|
0.607
|
NM_003632
|
CNTNAP1
|
contactin associated protein 1
|
|
chr10_+_111969988
|
0.607
|
NM_001008541
|
MXI1
|
MAX interactor 1
|
|
chr20_+_44034632
|
0.607
|
NM_001197139
NM_001048221
NM_001048222
NM_001197140
NM_018478
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr12_-_7261788
|
0.604
|
NM_016546
|
C1RL
|
complement component 1, r subcomponent-like
|
|
chr6_+_31623688
|
0.602
|
|
APOM
|
apolipoprotein M
|
|
chr17_-_7154994
|
0.600
|
NM_001143775
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1
|