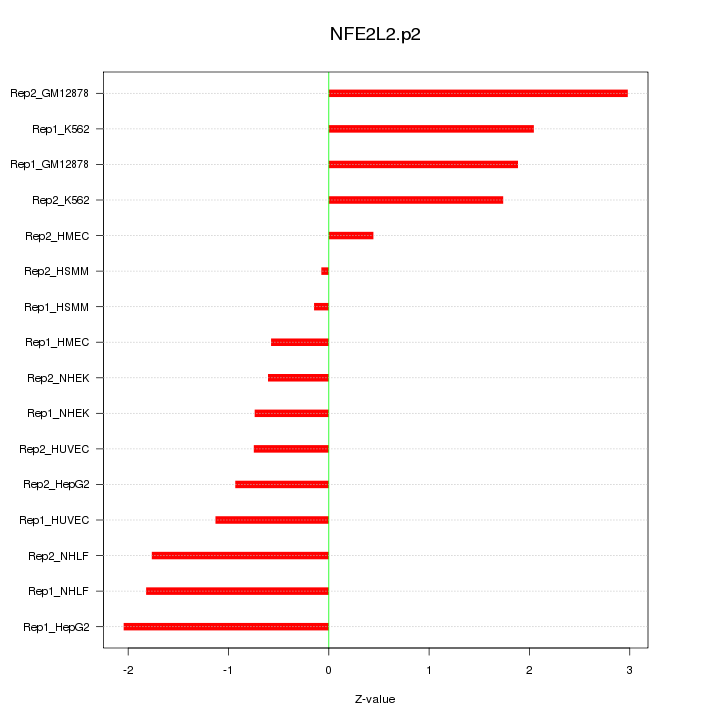
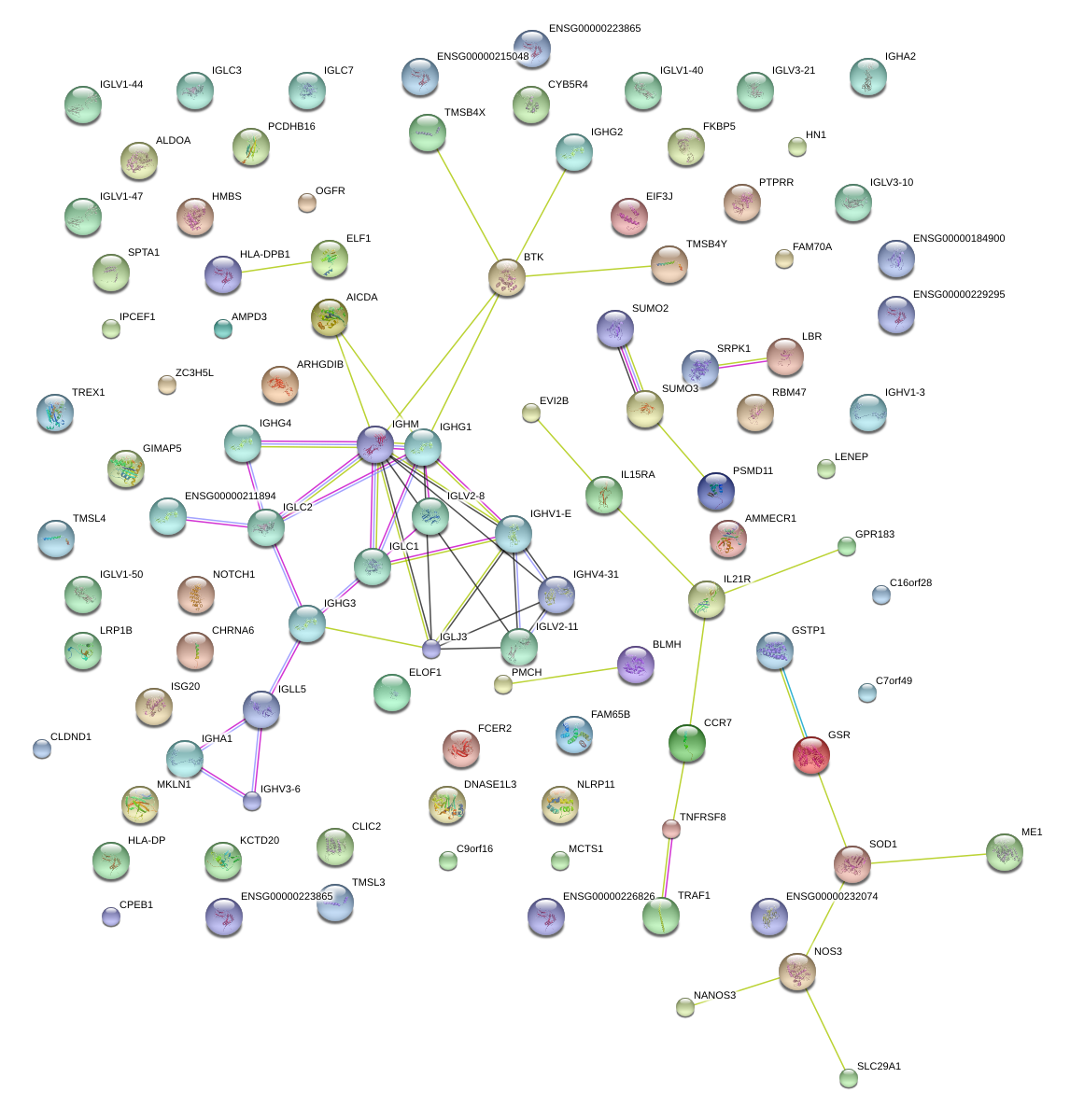
|
chr22_+_23010757
|
8.990
|
|
|
|
|
chrX_-_154563735
|
5.274
|
NM_001289
|
CLIC2
|
chloride intracellular channel 2
|
|
chr22_+_22569155
|
5.209
|
|
|
|
|
chr22_+_23040394
|
4.274
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chrX_-_100641143
|
4.102
|
NM_000061
|
BTK
|
Bruton agammaglobulinemia tyrosine kinase
|
|
chr22_+_22735134
|
3.992
|
|
CYAT1
|
immunoglobulin lambda light chain-like
|
|
chr22_+_22723972
|
3.675
|
|
|
|
|
chr19_-_7764283
|
3.126
|
NM_001207019
|
FCER2
|
Fc fragment of IgE, low affinity II, receptor for (CD23)
|
|
chr22_+_23229959
|
3.121
|
NM_001178126
|
IGLL5
IGLC1
|
immunoglobulin lambda-like polypeptide 5
immunoglobulin lambda constant 1 (Mcg marker)
|
|
chr22_+_22735207
|
3.090
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr22_+_22758538
|
3.082
|
|
|
|
|
chr22_+_22712091
|
3.079
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr22_+_23165273
|
3.045
|
|
|
|
|
chr9_-_123688352
|
3.015
|
|
TRAF1
|
TNF receptor-associated factor 1
|
|
chr17_+_30771520
|
2.967
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr11_+_67351276
|
2.645
|
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chr11_+_67351065
|
2.617
|
NM_000852
|
GSTP1
|
glutathione S-transferase pi 1
|
|
chrX_+_12993224
|
2.417
|
NM_021109
|
TMSB4X
|
thymosin beta 4, X-linked
|
|
chr22_+_22786283
|
2.415
|
|
|
|
|
chr13_-_99959649
|
2.332
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr17_-_38721701
|
2.180
|
NM_001838
|
CCR7
|
chemokine (C-C motif) receptor 7
|
|
chr17_+_30771494
|
2.097
|
NM_002815
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr7_-_134854225
|
2.021
|
NM_001243753
NM_001243755
|
C7orf49
|
chromosome 7 open reading frame 49
|
|
chr14_-_106733392
|
2.001
|
|
IGHG1
IGHG3
|
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr12_-_15114495
|
1.988
|
NM_001175
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta
|
|
chr17_+_30771522
|
1.977
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr17_+_30771532
|
1.959
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr17_-_28618836
|
1.953
|
|
BLMH
|
bleomycin hydrolase
|
|
chr17_+_30771485
|
1.941
|
|
PSMD11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11
|
|
chr6_+_84569427
|
1.915
|
|
CYB5R4
|
cytochrome b5 reductase 4
|
|
chr22_+_22676814
|
1.901
|
|
|
|
|
chr14_-_106967528
|
1.868
|
|
IGHA1
IGHA2
IGHG1
IGHM
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant mu
|
|
chr6_-_24877453
|
1.860
|
NM_015864
|
FAM65B
|
family with sequence similarity 65, member B
|
|
chr6_+_84569394
|
1.843
|
|
CYB5R4
|
cytochrome b5 reductase 4
|
|
chr12_-_8765438
|
1.819
|
NM_020661
|
AICDA
|
activation-induced cytidine deaminase
|
|
chr14_-_106539099
|
1.713
|
|
IGHA1
IGHG1
IGHG3
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
|
|
chr11_+_10476665
|
1.696
|
NM_001025389
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr14_-_106641585
|
1.685
|
|
IGHV4-31
IGHA1
IGHA2
IGHG1
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant alpha 2 (A2m marker)
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr17_-_29641087
|
1.683
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr22_+_23222897
|
1.676
|
|
CYAT1
|
immunoglobulin lambda light chain-like
|
|
chr11_+_118958696
|
1.659
|
NM_001024382
|
HMBS
|
hydroxymethylbilane synthase
|
|
chrX_-_109683457
|
1.645
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr1_-_158656487
|
1.595
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr6_+_84569364
|
1.500
|
NM_016230
|
CYB5R4
|
cytochrome b5 reductase 4
|
|
chr10_-_6019880
|
1.488
|
NM_001243539
|
IL15RA
|
interleukin 15 receptor, alpha
|
|
chr3_-_98241357
|
1.488
|
|
CLDND1
|
claudin domain containing 1
|
|
chr1_-_225616556
|
1.480
|
NM_194442
|
LBR
|
lamin B receptor
|
|
chr12_-_102591547
|
1.479
|
NM_002674
|
PMCH
|
pro-melanin-concentrating hormone
|
|
chr6_+_44195008
|
1.469
|
|
SLC29A1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr3_-_58200397
|
1.444
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr16_+_27438578
|
1.438
|
NM_021798
|
IL21R
|
interleukin 21 receptor
|
|
chr22_+_23154240
|
1.427
|
|
|
|
|
chr22_+_23134980
|
1.408
|
|
|
|
|
chr17_-_73150769
|
1.363
|
NM_001002032
NM_001002033
NM_016185
|
HN1
|
hematological and neurological expressed 1
|
|
chr15_+_89182038
|
1.357
|
NM_002201
|
ISG20
|
interferon stimulated exonuclease gene 20kDa
|
|
chr12_-_71182699
|
1.333
|
NM_001207015
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr1_+_154966061
|
1.308
|
NM_018655
|
LENEP
|
lens epithelial protein
|
|
chr9_+_130922471
|
1.306
|
NM_024112
|
C9orf16
|
chromosome 9 open reading frame 16
|
|
chr17_-_28619183
|
1.296
|
NM_000386
|
BLMH
|
bleomycin hydrolase
|
|
chr14_-_107170400
|
1.284
|
|
IGHV4-31
IGHV1-69
IGHA1
IGHG1
IGHG3
IGHM
|
immunoglobulin heavy variable 4-31
immunoglobulin heavy variable 1-69
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
immunoglobulin heavy constant gamma 3 (G3m marker)
immunoglobulin heavy constant mu
|
|
chr14_-_106926393
|
1.280
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chrX_+_119738542
|
1.269
|
NM_001137554
|
MCTS1
|
malignant T cell amplified sequence 1
|
|
chr7_+_150688143
|
1.252
|
NM_000603
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr22_+_22764094
|
1.237
|
|
|
|
|
chr10_-_6019454
|
1.233
|
NM_002189
NM_172200
|
IL15RA
|
interleukin 15 receptor, alpha
|
|
chr8_-_42623928
|
1.230
|
NM_001199279
NM_004198
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6
|
|
chr7_+_150434435
|
1.204
|
NM_018384
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr6_+_33043702
|
1.173
|
NM_002121
|
HLA-DPB1
|
major histocompatibility complex, class II, DP beta 1
|
|
chr6_-_35696359
|
1.146
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr4_-_40517913
|
1.142
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chrX_-_119445306
|
1.126
|
NM_001104544
NM_001104545
NM_017938
|
FAM70A
|
family with sequence similarity 70, member A
|
|
chr19_-_56348127
|
1.125
|
NM_145007
|
NLRP11
|
NLR family, pyrin domain containing 11
|
|
chr7_+_130794854
|
1.119
|
NM_001145354
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs
|
|
chr21_+_33031932
|
1.112
|
NM_000454
|
SOD1
|
superoxide dismutase 1, soluble
|
|
chr6_+_36410800
|
1.112
|
|
KCTD20
|
potassium channel tetramerisation domain containing 20
|
|
chr14_-_107122358
|
1.109
|
|
|
|
|
chr6_+_36410543
|
1.089
|
NM_173562
|
KCTD20
|
potassium channel tetramerisation domain containing 20
|
|
chr6_-_84140758
|
1.079
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr6_-_84140744
|
1.077
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr1_+_12123433
|
1.036
|
NM_001243
|
TNFRSF8
|
tumor necrosis factor receptor superfamily, member 8
|
|
chr3_+_48507209
|
1.030
|
NM_016381
NM_033629
|
TREX1
|
three prime repair exonuclease 1
|
|
chr16_+_30077120
|
1.024
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr6_-_35888816
|
1.018
|
|
SRPK1
|
SRSF protein kinase 1
|
|
chr15_+_44829373
|
1.017
|
|
EIF3J
|
eukaryotic translation initiation factor 3, subunit J
|
|
chr6_-_154677868
|
1.014
|
NM_001130700
NM_015553
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr8_-_30585220
|
0.987
|
|
GSR
|
glutathione reductase
|
|
chr15_-_83240499
|
0.980
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr21_-_46237882
|
0.977
|
|
SUMO3
|
SMT3 suppressor of mif two 3 homolog 3 (S. cerevisiae)
|
|
chr17_-_29641094
|
0.955
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr13_-_41593405
|
0.939
|
|
ELF1
|
E74-like factor 1 (ets domain transcription factor)
|
|
chr2_-_142889269
|
0.934
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr16_+_30077146
|
0.930
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr15_+_44829332
|
0.929
|
|
EIF3J
|
eukaryotic translation initiation factor 3, subunit J
|
|
chr16_-_1429612
|
0.915
|
NM_001193389
NM_023076
|
UNKL
|
unkempt homolog (Drosophila)-like
|
|
chr3_-_98241748
|
0.909
|
|
CLDND1
|
claudin domain containing 1
|
|
chrX_-_110513750
|
0.908
|
NM_014289
|
CAPN6
|
calpain 6
|
|
chr19_-_6670598
|
0.891
|
NM_003807
NM_172014
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14
|
|
chr15_+_44829265
|
0.883
|
NM_003758
|
EIF3J
|
eukaryotic translation initiation factor 3, subunit J
|
|
chr17_-_29641068
|
0.878
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr14_-_106774311
|
0.873
|
|
IGHA1
|
immunoglobulin heavy constant alpha 1
|
|
chr6_+_106546736
|
0.863
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr19_-_6670078
|
0.854
|
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14
|
|
chr16_+_30076993
|
0.847
|
NM_001243177
NM_184041
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr6_+_150070844
|
0.846
|
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase
|
|
chr22_-_19466725
|
0.844
|
NM_001035247
NM_005659
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast)
|
|
chr2_+_231921625
|
0.843
|
|
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1
|
|
chr10_-_76868853
|
0.843
|
NM_001007271
NM_001007272
NM_001007273
|
DUSP13
|
dual specificity phosphatase 13
|
|
chr3_+_48507621
|
0.832
|
|
TREX1
|
three prime repair exonuclease 1
|
|
chr11_-_3862183
|
0.824
|
NM_001665
|
RHOG
|
ras homolog gene family, member G (rho G)
|
|
chr6_+_150070818
|
0.809
|
NM_001252049
NM_001252050
NM_001252051
NM_001252052
NM_001252053
NM_005389
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase
|
|
chr22_-_19466621
|
0.805
|
|
UFD1L
|
ubiquitin fusion degradation 1 like (yeast)
|
|
chr6_+_32121228
|
0.802
|
NM_138717
|
PPT2
|
palmitoyl-protein thioesterase 2
|
|
chr19_-_2038937
|
0.801
|
|
MKNK2
|
MAP kinase interacting serine/threonine kinase 2
|
|
chr22_+_22735202
|
0.801
|
|
|
|
|
chr5_-_176936857
|
0.801
|
NM_024872
|
DOK3
|
docking protein 3
|
|
chr7_+_107204402
|
0.795
|
NM_181581
|
DUS4L
|
dihydrouridine synthase 4-like (S. cerevisiae)
|
|
chr6_-_35888889
|
0.793
|
|
SRPK1
|
SRSF protein kinase 1
|
|
chr19_-_6591012
|
0.792
|
|
CD70
|
CD70 molecule
|
|
chr22_+_23063107
|
0.788
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chrX_-_119445279
|
0.783
|
|
FAM70A
|
family with sequence similarity 70, member A
|
|
chr7_+_139528951
|
0.781
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chr11_-_3862137
|
0.780
|
|
RHOG
|
ras homolog gene family, member G (rho G)
|
|
chr6_-_35888937
|
0.771
|
NM_003137
|
SRPK1
|
SRSF protein kinase 1
|
|
chr1_-_150208441
|
0.770
|
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E
|
|
chr1_-_246580710
|
0.761
|
NM_022743
|
SMYD3
|
SET and MYND domain containing 3
|
|
chrX_-_64754594
|
0.754
|
|
LAS1L
|
LAS1-like (S. cerevisiae)
|
|
chr10_+_85899184
|
0.750
|
NM_014394
|
GHITM
|
growth hormone inducible transmembrane protein
|
|
chr17_+_43299277
|
0.749
|
NM_005892
|
FMNL1
|
formin-like 1
|
|
chr6_+_44214853
|
0.739
|
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1
|
|
chr12_-_57030083
|
0.733
|
NM_013449
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr10_+_121411086
|
0.730
|
|
BAG3
|
BCL2-associated athanogene 3
|
|
chrX_-_53350521
|
0.728
|
NM_001111125
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr6_+_150070856
|
0.725
|
|
PCMT1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase
|
|
chr12_+_64845839
|
0.724
|
NM_013254
|
TBK1
|
TANK-binding kinase 1
|
|
chr15_+_81517639
|
0.723
|
NM_172217
|
IL16
|
interleukin 16
|
|
chr2_-_233641225
|
0.722
|
NM_001172416
NM_001172417
NM_002242
|
KCNJ13
|
potassium inwardly-rectifying channel, subfamily J, member 13
|
|
chr7_+_148395886
|
0.716
|
NM_003592
|
CUL1
|
cullin 1
|
|
chr11_+_10477480
|
0.704
|
NM_001025390
|
AMPD3
|
adenosine monophosphate deaminase 3
|
|
chr1_+_233749749
|
0.701
|
NM_002245
|
KCNK1
|
potassium channel, subfamily K, member 1
|
|
chr1_-_109969044
|
0.701
|
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr7_+_148396016
|
0.698
|
|
CUL1
|
cullin 1
|
|
chr8_-_59571920
|
0.696
|
NM_001144772
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor
|
|
chr15_+_40733302
|
0.695
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr10_-_99052429
|
0.691
|
NM_001204300
NM_032900
|
ARHGAP19
|
Rho GTPase activating protein 19
|
|
chr6_+_44214786
|
0.689
|
NM_007355
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1
|
|
chr8_-_59572403
|
0.687
|
NM_003580
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor
|
|
chr1_-_150208462
|
0.681
|
NM_001136478
NM_030920
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E
|
|
chr1_-_109969058
|
0.676
|
NM_001199772
NM_002790
NM_001199773
NM_001199774
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr17_+_73598139
|
0.676
|
|
MYO15B
|
myosin XVB pseudogene
|
|
chr22_+_23029175
|
0.673
|
|
IGLV3-25
|
immunoglobulin lambda variable 3-25
|
|
chr2_-_220173693
|
0.667
|
NM_001199764
|
PTPRN
|
protein tyrosine phosphatase, receptor type, N
|
|
chrX_-_72434623
|
0.660
|
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr17_-_45899133
|
0.654
|
NM_145798
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr11_-_14541867
|
0.642
|
|
PSMA1
|
proteasome (prosome, macropain) subunit, alpha type, 1
|
|
chrX_-_64754637
|
0.636
|
NM_001170649
NM_001170650
NM_031206
|
LAS1L
|
LAS1-like (S. cerevisiae)
|
|
chr22_-_39268214
|
0.636
|
|
CBX6
|
chromobox homolog 6
|
|
chr11_-_47447457
|
0.635
|
|
PSMC3
|
proteasome (prosome, macropain) 26S subunit, ATPase, 3
|
|
chr4_-_76944585
|
0.627
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr21_-_33651324
|
0.617
|
NM_018944
|
MIS18A
|
MIS18 kinetochore protein homolog A (S. pombe)
|
|
chr14_+_103800678
|
0.614
|
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr1_+_14075861
|
0.614
|
NM_001007257
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr6_+_34206567
|
0.613
|
NM_145905
|
HMGA1
|
high mobility group AT-hook 1
|
|
chr12_-_76478737
|
0.607
|
NM_004537
NM_139207
|
NAP1L1
|
nucleosome assembly protein 1-like 1
|
|
chr16_+_90168671
|
0.607
|
|
FAM157C
|
family with sequence similarity 157, member C
|
|
chr7_+_148395995
|
0.605
|
|
CUL1
|
cullin 1
|
|
chr7_+_150434679
|
0.598
|
|
GIMAP5
|
GTPase, IMAP family member 5
|
|
chr2_-_89292396
|
0.595
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr1_-_109968951
|
0.594
|
|
PSMA5
|
proteasome (prosome, macropain) subunit, alpha type, 5
|
|
chr18_-_267987
|
0.592
|
NM_005131
|
THOC1
|
THO complex 1
|
|
chr3_+_133118789
|
0.587
|
NM_003571
|
BFSP2
|
beaded filament structural protein 2, phakinin
|
|
chr3_+_132373289
|
0.586
|
NM_198329
|
UBA5
|
ubiquitin-like modifier activating enzyme 5
|
|
chr12_+_56137063
|
0.580
|
NM_005811
|
GDF11
|
growth differentiation factor 11
|
|
chr8_-_59572385
|
0.575
|
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor
|
|
chr3_+_184038050
|
0.568
|
NM_004953
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr16_-_67517715
|
0.567
|
NM_001138
|
AGRP
|
agouti related protein homolog (mouse)
|
|
chrX_-_129244422
|
0.566
|
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr1_-_150208134
|
0.562
|
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E
|
|
chr17_-_45899118
|
0.561
|
|
OSBPL7
|
oxysterol binding protein-like 7
|
|
chr3_+_141043054
|
0.557
|
NM_001080412
|
ZBTB38
|
zinc finger and BTB domain containing 38
|
|
chr18_+_158480
|
0.554
|
NM_001037334
NM_005151
|
USP14
|
ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase)
|
|
chr2_-_214016332
|
0.551
|
NM_001079526
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr17_-_65362635
|
0.547
|
|
PSMD12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12
|
|
chr10_+_115312777
|
0.544
|
NM_004132
|
HABP2
|
hyaluronan binding protein 2
|
|
chr10_+_131934674
|
0.544
|
|
GLRX3
|
glutaredoxin 3
|
|
chrX_-_72434656
|
0.536
|
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr7_+_112063131
|
0.536
|
NM_001007245
NM_001197080
|
IFRD1
|
interferon-related developmental regulator 1
|
|
chr3_-_98241722
|
0.536
|
|
CLDND1
|
claudin domain containing 1
|
|
chr10_-_94333721
|
0.531
|
|
IDE
|
insulin-degrading enzyme
|
|
chr5_+_179125362
|
0.530
|
|
CANX
|
calnexin
|
|
chr6_-_84140651
|
0.530
|
|
ME1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr19_+_18699494
|
0.529
|
NM_001100418
NM_001100419
|
C19orf60
|
chromosome 19 open reading frame 60
|
|
chr2_+_231921608
|
0.528
|
|
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1
|
|
chr3_+_184038445
|
0.527
|
|
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1
|
|
chr1_+_114493766
|
0.525
|
NM_198269
|
HIPK1
|
homeodomain interacting protein kinase 1
|
|
chr17_+_45286427
|
0.522
|
NM_001002841
|
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic
|
|
chr14_-_23284565
|
0.520
|
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7
|
|
chr1_+_14075931
|
0.517
|
|
PRDM2
|
PR domain containing 2, with ZNF domain
|
|
chr6_-_33548017
|
0.515
|
NM_001188
|
BAK1
|
BCL2-antagonist/killer 1
|
|
chr12_+_50794591
|
0.513
|
NM_001170803
NM_001170804
NM_001170808
NM_052879
NM_199188
NM_199190
|
LARP4
|
La ribonucleoprotein domain family, member 4
|
|
chr11_-_14541886
|
0.512
|
NM_001143937
NM_002786
|
PSMA1
|
proteasome (prosome, macropain) subunit, alpha type, 1
|